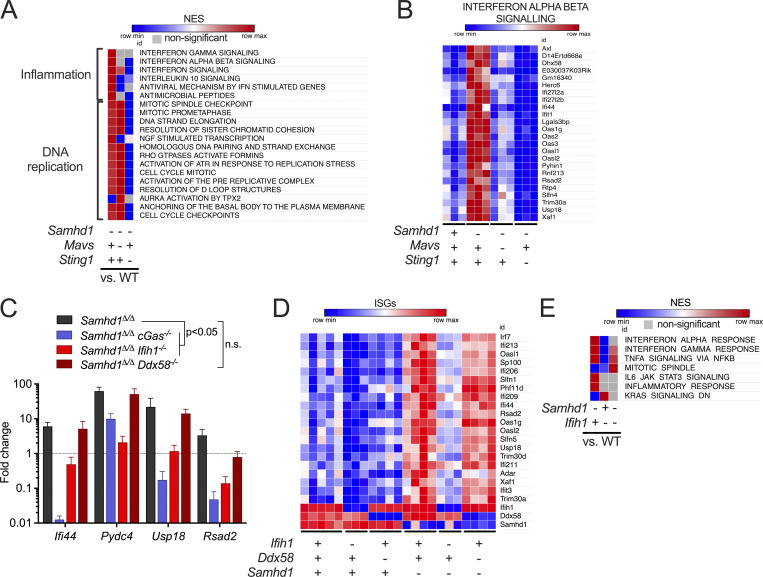
Figure 4.
MDA5 drives spontaneous IFN production in Samhd1Δ/Δ mice. For the whole figure − = homozygous null, + = homozygous WT. (A) Enrichment of reactome gene sets (MSigDB) in the transcriptome of peritoneal macrophages from mutant mice compared with littermate WT controls of Samhd1Δ/Δ mice. (B) Normalized read counts for the indicated ISG transcripts from the analysis shown in A. (C) Relative transcript levels of the indicated ISGs measured by qRT-PCR in post-replicative senescence Samhd1Δ/Δ MEFs with additional CRISPR-mediated inactivation of the genes cGas (n = 4), Ifih1 (n = 3), and Ddx58 (n = 2). Data from two independent experiments were pooled and displayed as fold change compared to the mean of Samhd1+/+ MEFs (multiple t tests, summary of results is shown with P < 0.05 as lowest significance level). (D) Differentially expressed (padj<0.1) ISG in transcriptomes of peritoneal macrophages from mutant mice compared with littermate WT controls of Samhd1Δ/Δ mice. (E) Enrichment of reactome gene sets (MSigDB) of another independently generated transcriptome using peritoneal macrophages from Samhd1Δ/ΔIfih1+/+, Samhd1+/+Ifih1−/−, and Samhd1Δ/ΔIfih1−/− compared with littermate Samhd1+/+Ifih1+/+control mice. NES, normalized enrichment score.