Figure 1. Cdx2-induced ESCs self-assemble with Gata4-induced ESCs and ESCs into post-implantation-like mouse embryoids.
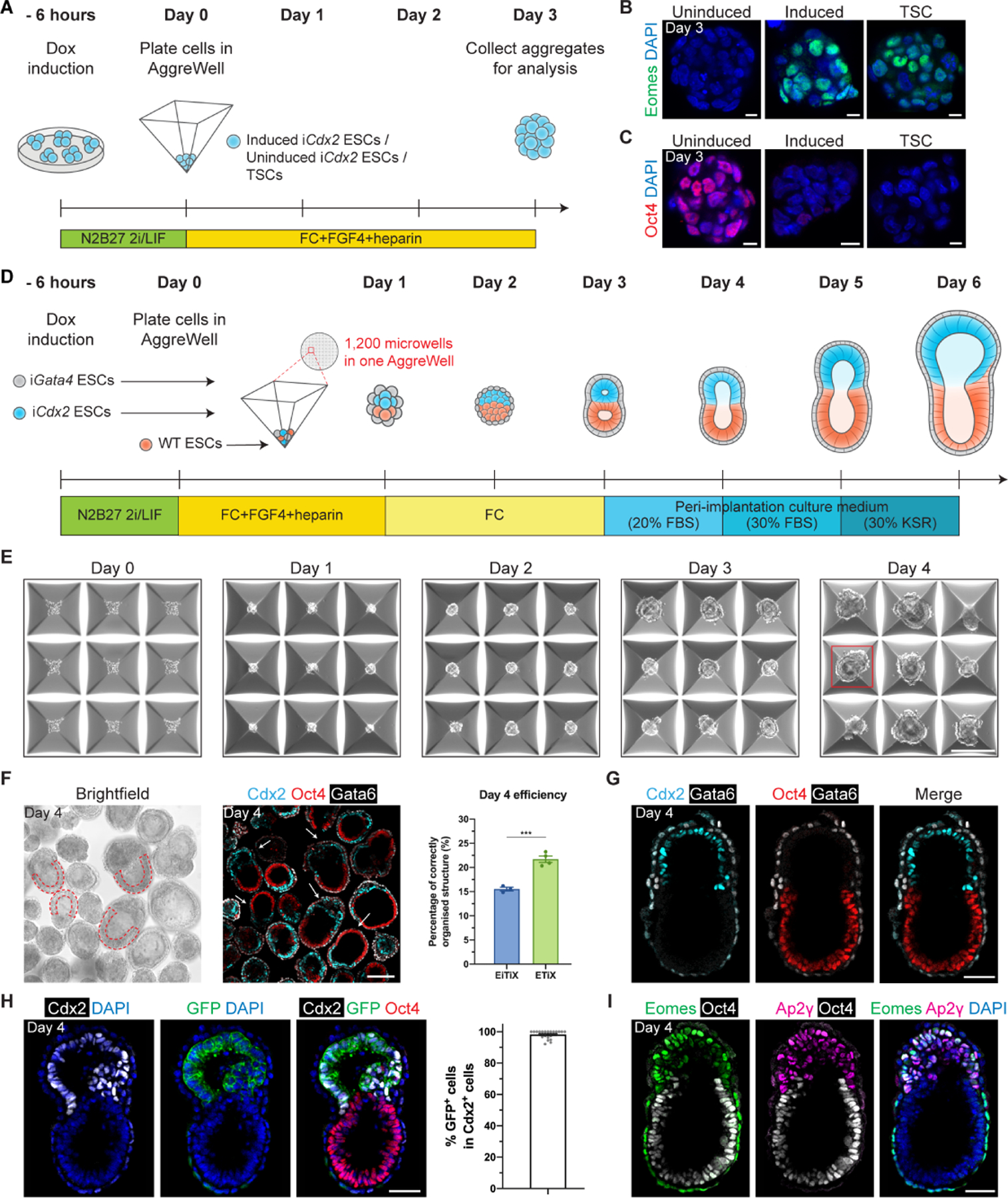
(A) Schematic of the formation of cell aggregates in AggreWells. Day 3 iCdx2 ESC aggregates show elevated Eomes expression (B) and downregulated Oct4 (C) upon induction of Cdx2. (D) Schematic of EiTiX-embryoid generation. FBS: fetal bovine serum, KSR: knockout serum replacement. (E) Representative brightfield images of structures developing in AggreWells from Day 0 to Day 4. A structure resembling the early post-implantation mouse embryo can be seen on Day 4 in the well outlined in red. (F) All structures in the combined microwells from one AggreWell were collected at Day 4 and stained to reveal Cdx2 (cyan), Oct4 (red) and Gata6 (white). Arrows indicate structures considered to exhibit correct organisation. Such cylindrical structures with two cellular compartments and an epithelialised EPI-like cell layer (red dashed outline) were selected under brightfield microscopy. The efficiency of obtaining organised structures in EiTiX-embryoid system and ETiX-embryoid system is shown. Efficiency of ETiX-embryoid is taken from our previous publication (referring to these embryoids as iETX-embryoids, Amadei et al., 2021). (G) Day 4 EiTiX-embryoids stained to reveal Cdx2 (cyan), Oct4 (red) and Gata6 (white). (H) Day 4 EiTiX-embryoid stained to reveal Cdx2 (white), GFP (green) and Oct4 (red). The percentage of the Cdx2-positive cells that are also GFP-positive is shown. n = 49 structures. (I) Day 4 EiTiX-embryoids stained to reveal Eomes (green), Ap2γ (magenta) and Oct4 (white). n = 35/35 structures are positive for both Eomes and Ap2γ. All experiments were performed minimum 3 times. Scale bars: 10μm, (B-C); 150μm, (E-F); 50μm, (G-I). See also Figure S1.
