Figure 2. EiTiX-embryoids establish an anterior-posterior axis and undergo gastrulation.
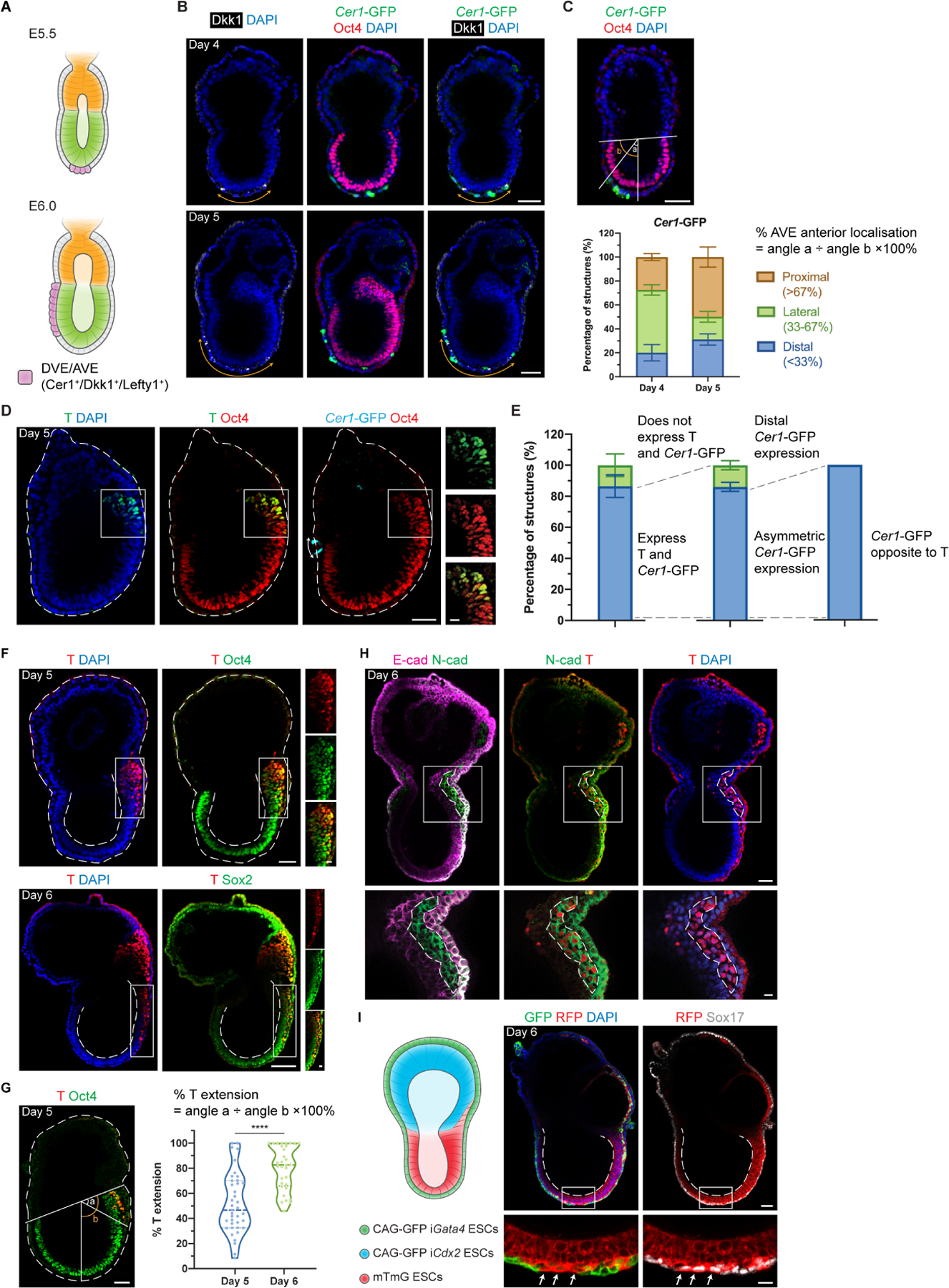
(A) Schematic showing the position of DVE/AVE in E5.5 and E6.5 mouse embryos. (B) Day 4 and Day 5 EiTiX-embryoids stained to reveal Cer1-GFP (green), Oct4 (red) and Dkk1 (white). Orange double-headed arrows indicate Dkk1-positive domains. (C) Localisation of Cer1-GFP in Day 4 and Day 5 EiTiX-embryoids (See Materials and Methods for quantification method). n = 57 Day 4 structures; 45 Day 5 structures. (D) Day 5 EiTiX-embryoids stained to reveal T (green), Oct4 (red) and Cer1-GFP (cyan). Double-headed arrow, Cer1-GFP-expressing domain; white box outline, T- and Oct4-positive domain. (E) Percentages of Day 5 EiTiX-embryoids showing 1) expression of both Cer1-GFP and T in same structure, 2) asymmetric Cer1-GFP expression, and 3) T expression on the opposite side from Cer1-GFP. n = 42 structures. (F) Day 5 and Day 6 EiTiX-embryoids stained to reveal T (red) and Oct4 (green) or Sox2 (green). White boxes enclose T-positive domain while dotted lines outline the structure and the lumen of ES compartment. n = 39/42 Day 5 structures and 32/32 Day 6 structures. (G) Percentage of T extension in Day 5 and Day 6 EiTiX-embryoids. See Materials and Methods for quantification method. n = 39 Day 5 structures and 32 Day 6 structures. ****p < 0.0001. (H) Day 6 EiTiX-embryoid stained to reveal E-cadherin (magenta), N-cadherin (green) and T (red). Dotted line indicates T- and N-cadherin-positive domain. n = 14/21 structures with N-cadherin upregulation and E-cadherin downregulation from 4 experiments. (I) Schematic of EiTiX-embryoids using CAG-GFP iGata4 ESCs (with membrane GFP) and mTmG ESCs (with membrane tdTomato) to construct the VE-like layer and EPI-like compartment, respectively. Day 6 EiTiX-embryoid stained to reveal GFP (green), RFP (red) and Sox17 (white). Dotted line indicates the lumen of ES compartment while arrows mark definitive endoderm-like cells intercalated into the VE-like layer. n = 8/12 structures. All experiments were performed minimum 3 times. Scale bars: 50μm; 15μm (zoomed). See also Figure S2.
