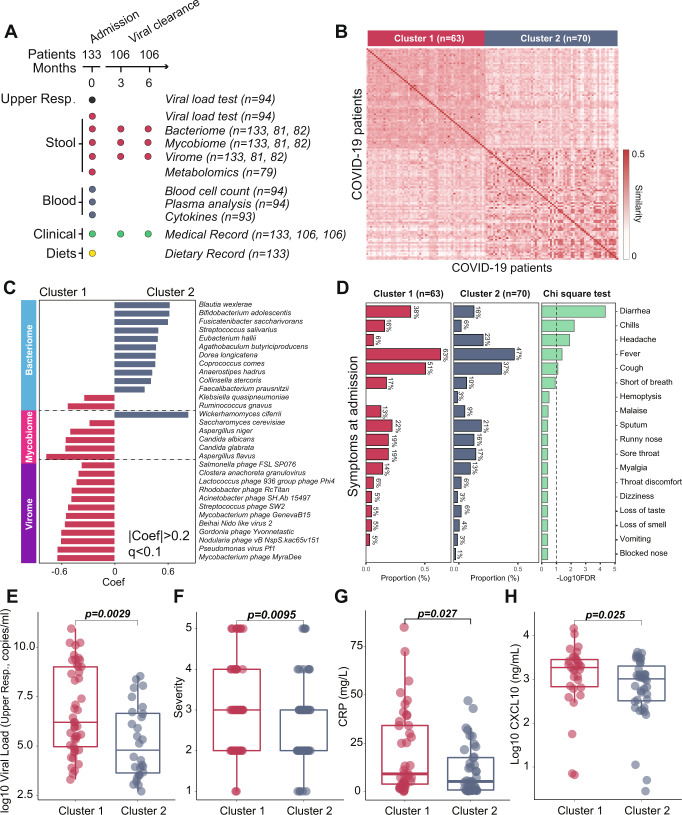
Fig. 2. Integration of gut multi-biome data through weighted similarity network fusion (WSNF) approach.
A Schematic overview of the study design, depicting the total number of samples and participants from whom data were available. B Heatmap illustrating pairwise patient WSNF similarity scores stratified by spectral clustering (Cluster 1, n = 63; Cluster 2, n = 70) according to integrated multi-biome profiles, derived from n = 133 biologically independent samples. C MaAslin2 analysis of observed clusters illustrating discriminant taxa at baseline (FDR-adjusted q < 0.1). D Symptoms of COVID-19 patients between two identified patient clusters. The proportion of diarrhea, chills, headache, fever, and cough in Cluster 1 were significantly higher than in Cluster 2 (Cluster 1, n = 63; Cluster 2, n = 70) (Chi-square test with one degree of freedom, Benjamini–Hochberg correction, p < 0.05; q < 0.1). E Comparison of viral load (copies/mL). F Severity of disease. G C-reactive protein (CRP) concentration. H CXCL levels in two identified patient clusters. In E–H, the two-sided Wilcoxon rank-sum test was used to check the differences between the two clusters. Boxplot lower and upper hinges correspond to the first and third quartiles, upper and lower whiskers represent the highest and lowest values within 1.5 times the interquartile range, and the horizontal line represents the median.