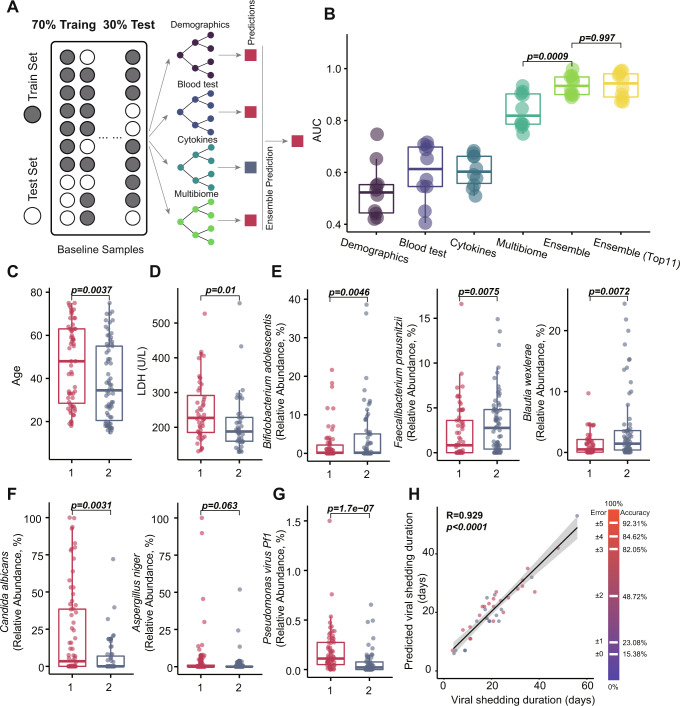
Fig. 4. Random forest classifier model trained on multi-biome and clinical data can predict the duration of viral shedding for individual COVID-19 patients.
A The input data is a vector with four components: demographics, blood tests, cytokines, and gut multi-biome profiles. To estimate model accuracies, a train-test sample split of 70% for training and 30% for testing was utilized. The testing data were then used to estimate the accuracy of the random forest model. B Box-and-whisker plot displaying the distribution of AUC scores for the cross-validation on the training set and the AUC scores for single measurements taken on the test set, obtained by random forest classification. Differences between groups were evaluated by the two-sided Wilcoxon rank-sum test. C–G Top features contribute to differentiating clusters (Cluster 1, n = 63; Cluster 2, n = 70) in the random forest models. In C–G, the two-sided Wilcoxon test was used to check the differences between the two clusters. H Integration of multi-biome and clinical data for predicting the duration of viral shedding of SARS-CoV-2. The predicted positive time was paired with the real positive time for accuracy evaluation, and the accuracy was calculated at different error levels from ±0 to ±5 days. Error bands reflect the 95% CI. R and P values were calculated by two-sided Spearman Correlation, p < 0.0001. In B–G, the horizontal line in the boxplot indicates the median value. Box plots lower and upper hinges correspond to the first and third quartiles and upper and lower whiskers represent the highest and lowest values within 1.5 times the interquartile range.