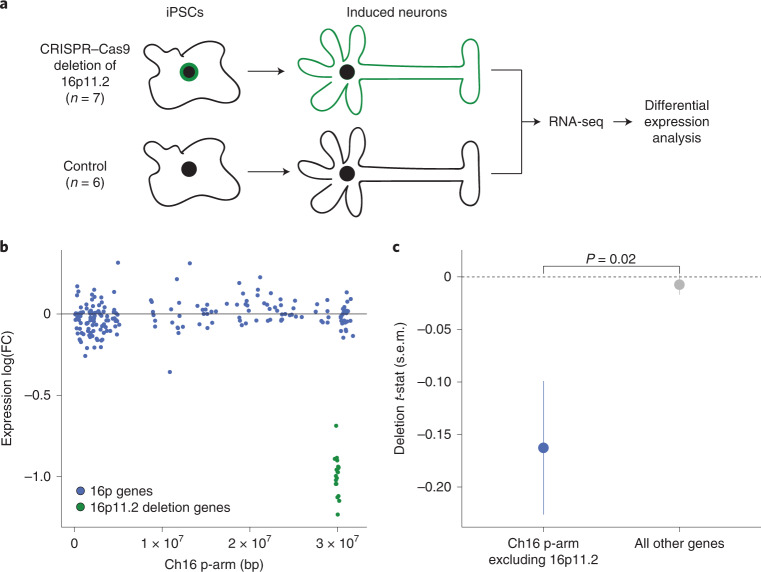
Fig. 2. In vitro deletion of 16p11.2 causes decreased average expression of neuronally expressed 16p genes.
a, Experimental design of 16p11.2 in vitro deletion resource. The iPSCs undergo CRISPR–Cas9-mediated deletion of the 16p11.2 CNV locus, differentiation into induced neurons and transcriptome profiling with RNA-seq (n = 7 biological replicates). Differential expression analysis compares these samples with controls (n = 6 biological replicates) without deletion of the locus. b, Differential expression of neuronally expressed genes on 16p (n = 200 genes) after deletion of the 16p11.2 locus (neuronally expressed is defined as above-median normalized expression level of genes over all samples in analysis of induced neurons). FC, fold-change. Genes in the deletion region ±0.1 Mb are green, whereas all other genes on 16p are in blue. The y axis is the log2(fold-change) per gene. c, The 16p11.2 deletion causing decreased expression of neuronally expressed genes on 16p (n = 200 genes), but not of all other neuronally expressed genes in the genome (n = 8,533 genes). Point estimates are of mean differential expression t-statistic for the group of genes ± s.e.m. The P value is from two-sided, two-sample Student’s t-test comparing groups.