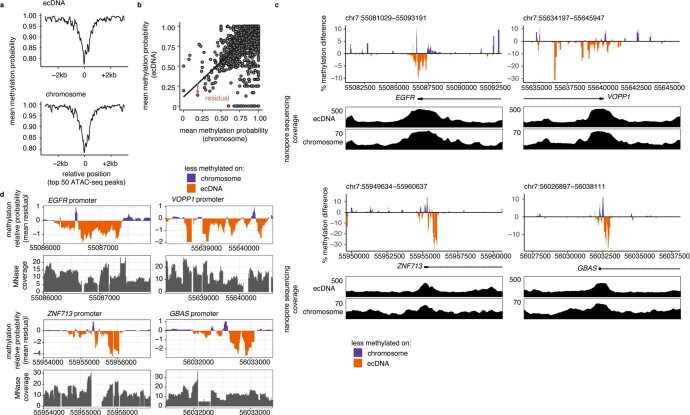
Extended Data Fig. 5. Quantification of 5mC-CpG methylation probability of ecDNA and the native chromosomal locus.
(a) Aggregated CpG methylation probability of ecDNA and chromosomal DNA at the top 50 ATAC-seq peaks with highest coverage in the amplified region. Mean methylation frequencies were calculated in 100-bp windows sliding every 10 bp. (b) Linear regression model of mean methylation probabilities of ecDNA vs chromosomal DNA. Mean methylation probabilities were calculated in 100-bp windows sliding every 10 bp in the ecDNA-amplified region. Each point represents a window mean. Brown arrow demonstrates the standardized residual of a data point from the regression line. (c) Relative CpG methylation of ecDNA compared to the chromosomal locus in differential regions shown as absolute differences in methylation frequencies. Regions shown correspond to differentially methylated regions in Fig. 4d,e. Mean methylation frequencies were calculated in 100-bp windows sliding every 10 bp. Normalized sequencing coverage tracks are shown on the bottom of each plot. (d) Relative CpG methylation of ecDNA compared to the chromosomal locus and nucleosome positioning by MNase-seq, zooming into indicated gene promoters. Regions shown correspond to differentially methylated regions in Fig. 4d,e. Mean methylation frequencies and MNase-seq coverage were calculated in 100-bp windows sliding every 10 bp. Relative frequencies were quantified from standardized residuals for a linear regression model for mean frequencies on ecDNA vs chromosomal DNA (Methods).