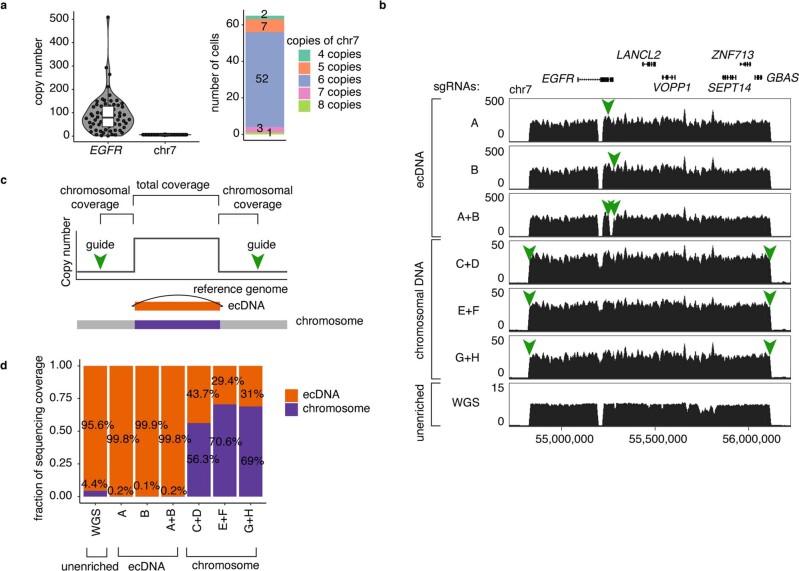
Extended Data Fig. 2. Enrichment of circular ecDNA by CRISPR-CATCH.
(a) Left: quantification of EGFR and chromosome 7 copy numbers in GBM39 cells using DNA FISH on metaphase spreads (n = 65 cells; box center line, median; box limits, upper and lower quartiles; box whiskers, 1.5× interquartile range). Right: number of GBM39 cells with 4, 5, 6, 7, or 8 copies of chromosome 7. An example FISH image is shown in Fig. 1b. (b) Full sequencing tracks showing coverage for isolated ecDNA and its chromosomal locus at the EGFR amplified region compared to WGS. Zoomed-in tracks are shown in Fig. 1f. Orange arrows indicate locations of sgRNA targets. (c) Chromosomal overhangs from chromosome-targeting guides (guides C-H) outside of the ecDNA-amplified region were used for calculating sequencing coverage of the chromosomal allele. The mean coverage of the 5’ and 3’ chromosomal overhangs was calculated. The coverage of ecDNA alleles was calculated by subtracting chromosomal coverage from total coverage in the ecDNA-amplified region. (d) Relative sequencing coverage of chromosomal DNA and ecDNA alleles in WGS or CRISPR-CATCH samples.