Figure 4:
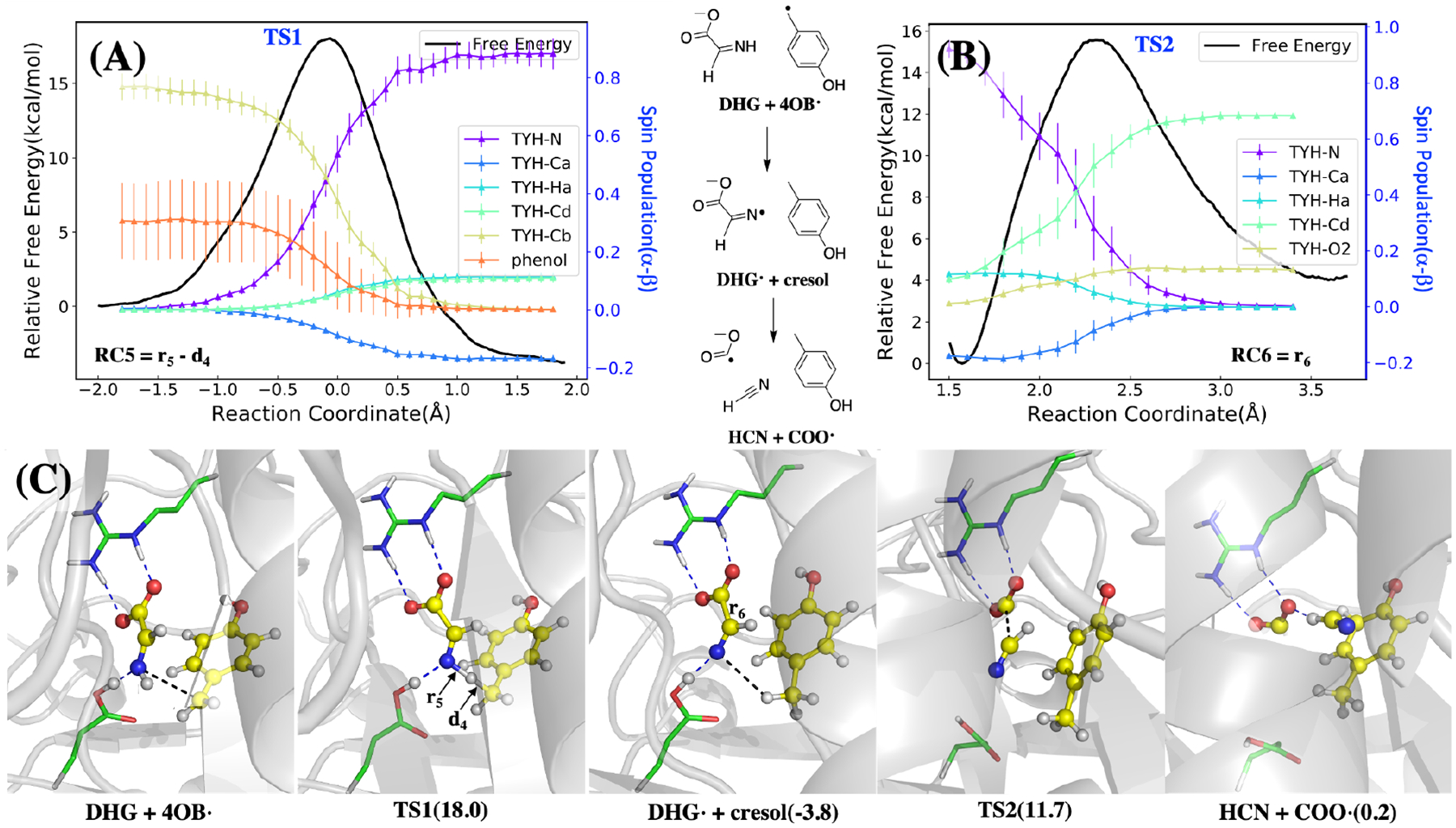
The calculated mechanism of DHG radical formation and decomposition, starting with H• abstraction from the DHG nitrogen, followed by decomposition of DHG radical to COO•− and HCN. Top: Free energy profile (A) and spin populations profiles (B) along the umbrella sampling reaction coordinates. The reaction coordinate of the DHG radical formation (top left) is defined as r5 − d4 where distances are indicated in the bottom left panel, and for the DHG radical decomposition it is defined as r6 shown in the bottom panel. The energy curves are both colored in black with the y-axis on the left, while the spin density profiles are shown for selected atoms in color with the y-axis are shown on the right. Bottom (C): Key structures during these two reactions with hydrogen bonds shown in blue dotted lines and the distances of the reaction coordinate shown in black dotted lines. As the structures shown in the bottom, the radical transfers from the 4-OB• to the nitrogen in the DHG, which helps the decomposition of the DHG into COO• and HCN. The barriers of these two steps are about 18 and 15.5 kcal/mol respectively. The radical transfer is exothermic while the DHG decomposition is endothermic and makes these two reactions overall barely absorb energy.
