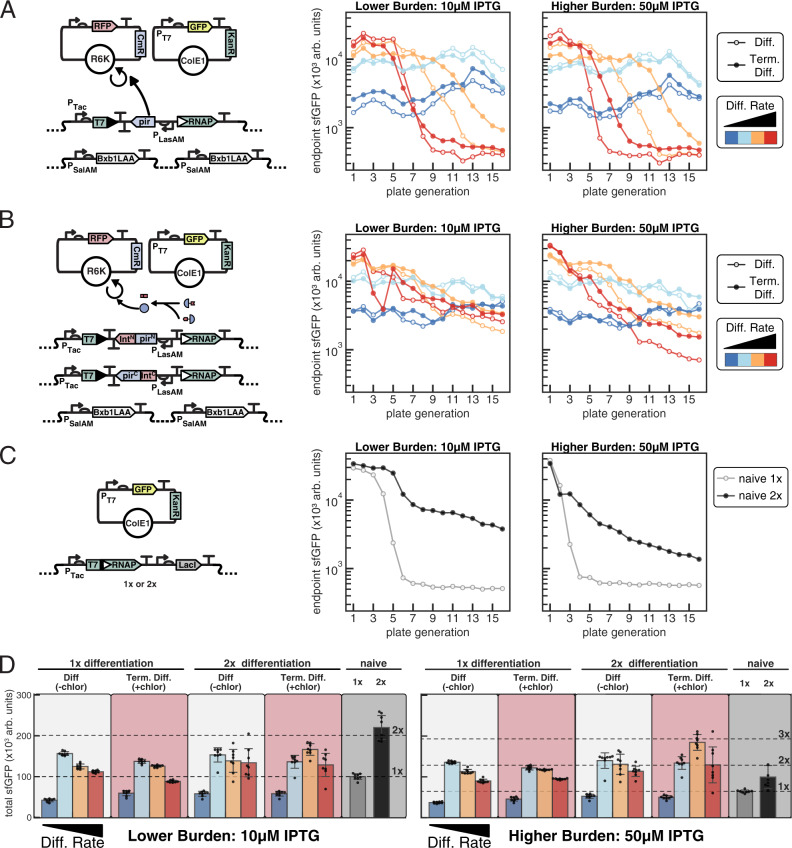
Fig. 2. Assessing the evolutionary stability of burdensome T7 RNAP driven expression from a high copy ColE1 KanR plasmid.
A–C 8 independent transformants were outgrown for 8 h in LB media with appropriate antibiotics and inducers before 50× dilution into experimental conditions. Cells were grown in 96 well plates in 0.3 mL, diluted 50× every 8 h for 16 total growths, and 50 μL endpoint samples taken to measure OD700, sfGFP (485/515 nm), and mScarlet (565/595 nM) fluorescence in 384 well Matriplates. Average endpoint sfGFP production (n = 8 biological replicates) plotted for each condition. A, B 1x differentiation and 2x differentiation. Each differentiation cassette additionally encodes NahRAM, LasRAM, and LacIAM (Supplementary Fig. 5). Cells were co-transformed with ColE1-KanR-PT7 GFP and R6K-CmR-mScarlet and plated on LB + kan/chlor/30 nM Las-AHL. Colonies were outgrown in LB + kan/chlor/10 nM Las-AHL before 50× dilution into experimental conditions in LB kan with chlor (terminal differentiation, filled circles) or without chlor (open circles) with varying concentrations of salicylate (10, 15, 20, 30 μΜ: dark blue, light blue, orange, red, respectively) and IPTG (10, 50 μΜ). C Cells with one (1x naive, grey) or two (2x naive, black) copies of genomically integrated T7 RNAP were transformed with ColE1-KanR-PT7 GFP, plated on LB + kan, and outgrown in LB + kan before dilution into experimental conditions. D Mean total cumulative production ± SD (n = 8 biological replicates) after 16 growths for all strains in all conditions shown in A-C. Dashed lines show 1×, 2×, 3× the average total GFP production of 1x naive. Source data are provided as a Source Data file.