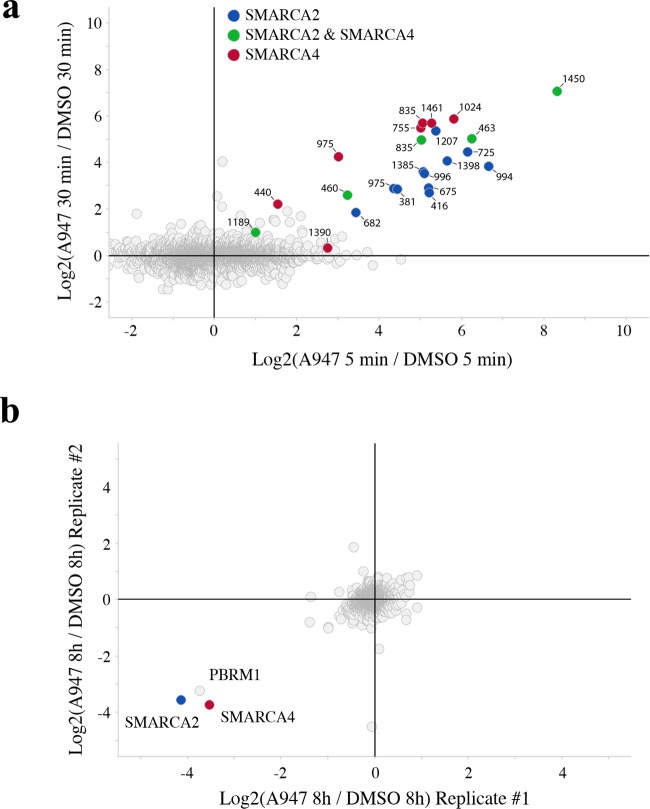
Fig. 2. A947-mediated ubiquitination and degradation of SMARCA2/4.
a Global ubiquitylome changes, as assessed by di-glycine reminant mass spectrometry profiling, in SW1573 cells following treatment with 500 nM A947 for 5 and 30 min. n = 5720 unique ubiquitinated peptides were identified and presented as log2 fold change relative to DMSO control cultures. Positions of ubiquitinated lysines in peptides that uniquely map to SMARCA2 (UniProtKB P-51531-1) or SMARCA4 (UniProtKB P-51532-2) are highlighted in blue and red, respectively. Peptides shared between SMARCA2/4 are shown in green, with ubiquitinated lysines mapped to the SMARCA2 sequence. b Global proteome assessed by mass spectrometry following 8 h treatment with A947 (100 nM) in SW1573 cells. Data are presented as a log2 fold change in the abundance of the respective proteins in two biological replicates. ~8900 and ~8400 proteins were quantified in replicate 1 and 2, respectively.