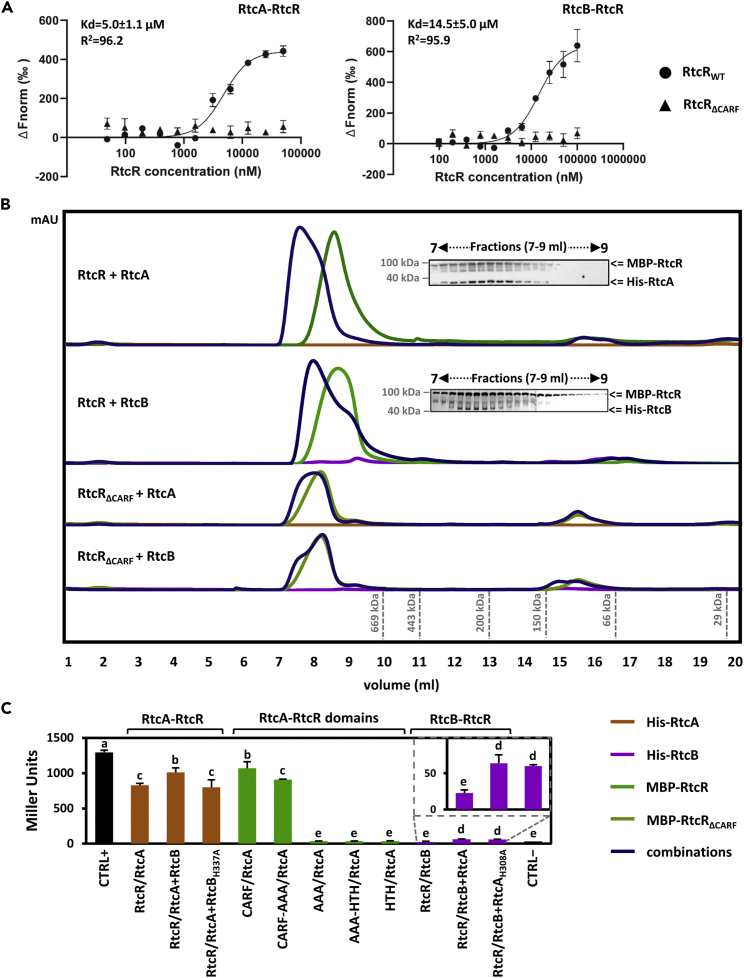
Figure 2.
The RtcRAB proteins interact via the RtcR CARF domain
(A and B) Rtc proteins were overexpressed and purified (Figure S2A), and their physical interactions were investigated using (A) MST and (B) gel filtration chromatography. (A) Changes in thermophoresis (y axis of graph) of fluorescently labeled RtcRWT and RtcRΔCARF was used to quantify their binding to RtcA or RtcB in titration experiments. RtcRWT, but not RtcRΔCARF, interacts in vitro with RtcA (left) and RtcB (right), as shown by MST. The dissociation constant (Kd) of RtcA and RtcB with RtcR is in the μM range. (B) Alterations in the fractionation and apparent molecular weight of RtcRWT and RtcRΔCARF were used to show their complex formation with RtcA or RtcB. The peak of MBP-RtcR, but not MBP-RtcRΔCARF, potentially forming a hexamer, shifts towards a higher molecular weight in the presence of His-RtcA and His-RtcB, as shown by gel filtration chromatography. Co-localization of MBP-RtcR with His-RtcA and His-RtcB was confirmed with SDS-PAGE and immunoblotting. The proteins were present at equimolar concentrations (20 μM).
(C) Pairwise combinations of N-terminal and C-terminal T18/T25 fusions between full length or truncated Rtc proteins (Figures S2B–S2D) were assessed for in vivo physical interactions as indicated by elevated β-galactosidase activity in bacterial 2-hybrid assays. RtcR interacts in vivo with RtcA via its CARF domain, as shown by bacterial two-hybrid analysis (N = 3). In trans RtcA or RtcB influences the interactions of RtcR with respectively RtcB and RtcA, as shown by bacterial three-hybrid analysis (N = 3). Data are shown as mean and error bars represent standard deviation from the mean. N represents total number of independent biological replicates, with 3 technical replicates each. ANOVA ∗ p-value < 0.05; ∗∗ p-value < 0.01; ∗∗∗ p-value < 0.001; ∗∗∗∗ p-value < 0.0001.