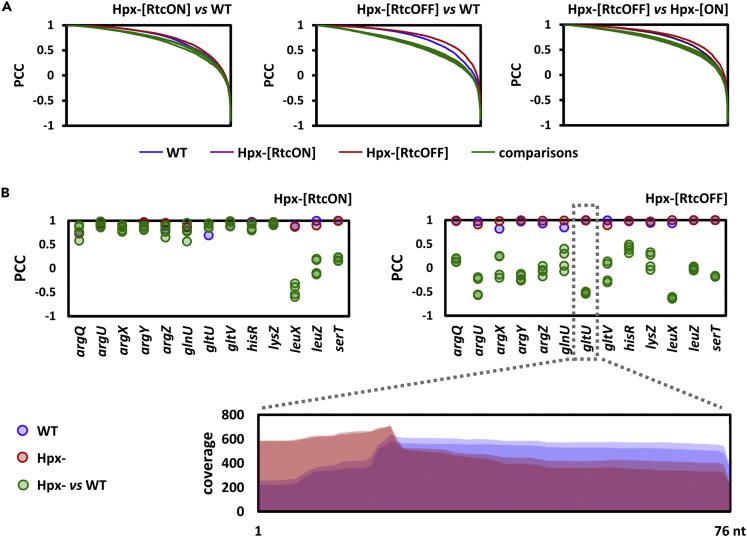
Figure 6.
tRNAs are cleaved under oxidative stress
Damaged RNAs in transcriptomes of E. coli Hpx-[RtcON] and Hpx-[RtcOFF] cells were identified by pairwise calculation of the Pearson’s Correlation Coefficient (PCC) for each individual gene.
(A) Globally, more transcripts are damaged in Hpx-[RtcOFF] than Hpx-[RtcON] cells compared to wild-type cells 8 h post-inoculation, as illustrated by plotting PCC values of all genes in descending order from high to low.
(B) More tRNAs are damaged in Hpx-[RtcOFF] (top right) than Hpx-[RtcON] (top left) compared to wild-type cells, as illustrated by plotting PCC values of pairwise comparisons for selected tRNAs. Distribution of all reads mapped to gltU in Hpx-[RtcOFF] and wild-type cells reveals no obvious cleavage sites (bottom).