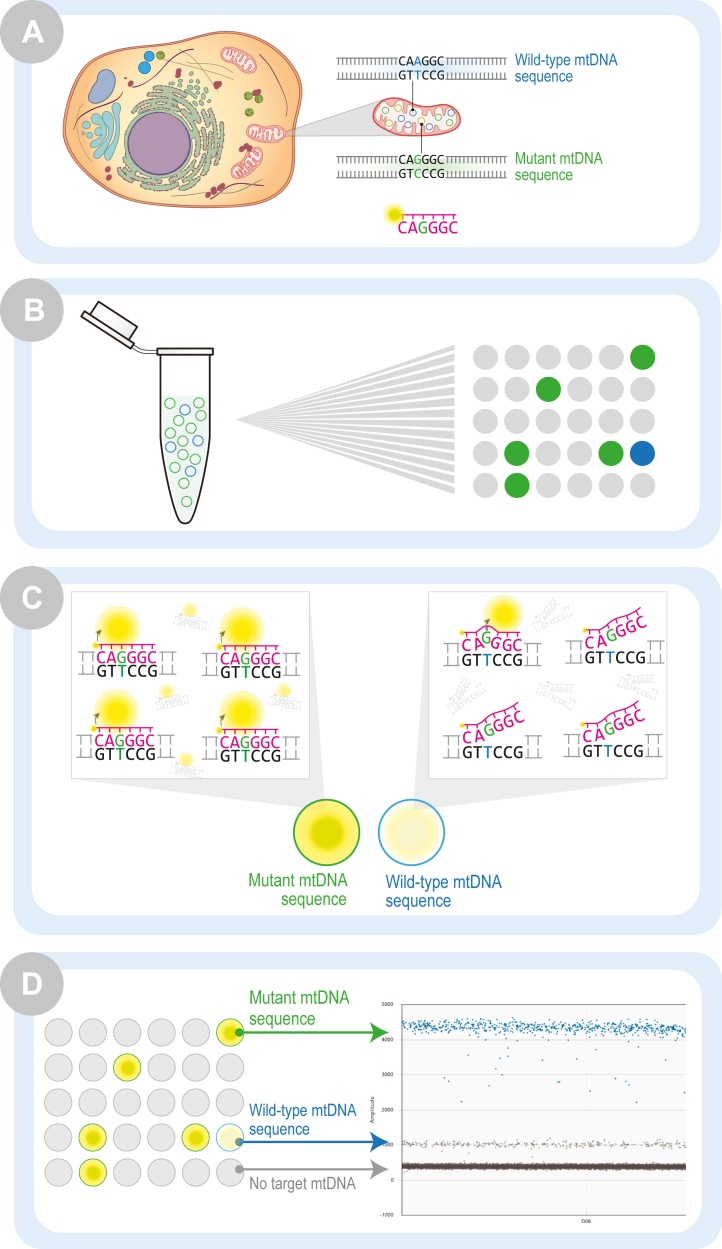
Figure 1.
Quantification of mtDNA heteroplasmy using a single dPCR probe. A, mtDNA mutations are commonly heteroplasmic, with both mutant and WT mtDNA present in the same cell. The abundance of each haplotype can be quantified using a fluorescent-labeled dPCR probe specific to one of the haplotypes, in this case the mutant mtDNA sequence. B, a bulk reaction mixture containing both mtDNA sequences is fractionated into partitions. Some partitions will contain WT mtDNA, some will contain mutant mtDNA, and some will contain no target DNA. PCR is carried out within each partition. C, when the mutant mtDNA sequence is present, efficient probe binding results in abundant fluorescent signal. When the WT mtDNA sequence is present, less efficient probe binding results in diminished, yet still detectable fluorescent signal. D, each partition is analyzed as either positive or negative for the fluorescent signal. Partitions are graphically distributed based on the abundance of fluorescent signal present in the partition where more fluorescent signal produces a higher amplitude partition population. mtDNA, mitochondrial DNA; dPCR, digital PCR.