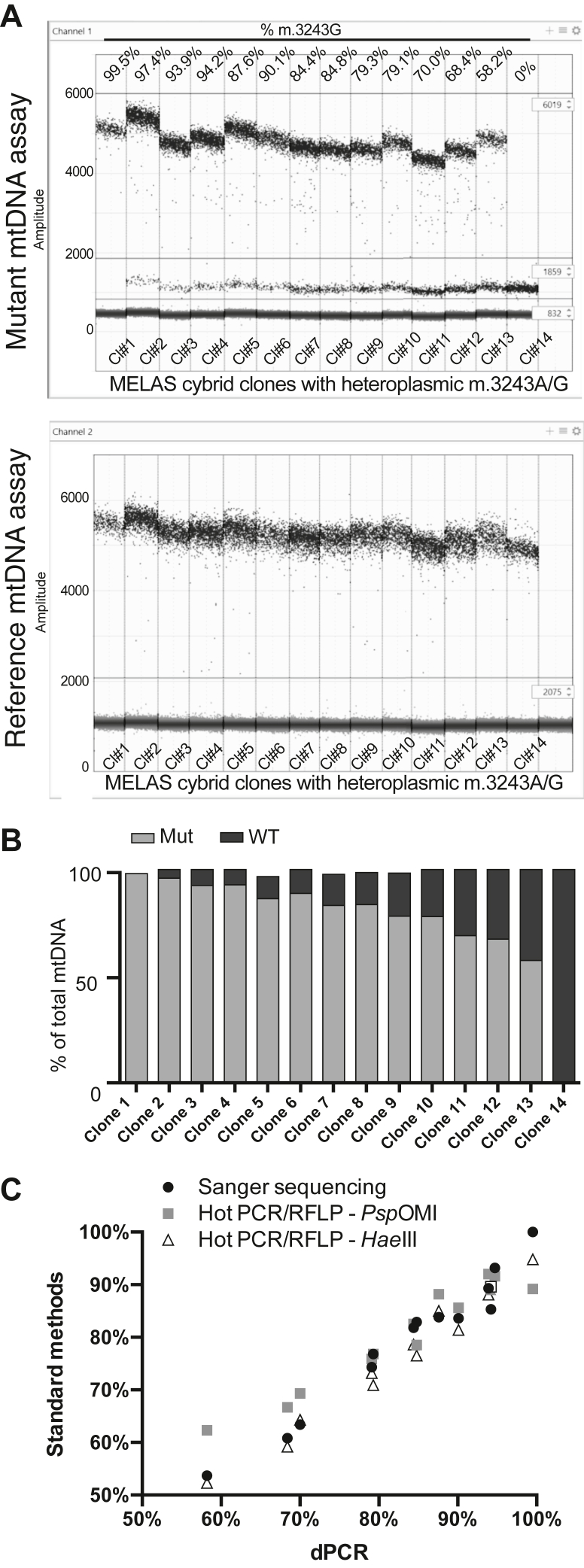
Figure 3.
dPCR can be used to quantify heteroplasmy in cellular DNA samples. A, duplex droplet dPCR plots for the mutant mtDNA assay (top) and reference mtDNA assay (bottom) using various m.3243A > G heteroplasmic cell lines (clones #1–14). The mutant mtDNA assay used a FAM-labeled probe, while the reference mtDNA assay used a HEX-labeled probe. Horizontal lines are drawn to differentiate the distinct droplet populations in the mutant mtDNA assay (high-amplitude positive, low-amplitude positive, and negative) and reference mtDNA assay (positive and negative). The calculated percentage of mutant mtDNA is displayed above each sample. B, heteroplasmy quantification of various m.3243A > G heteroplasmic cell lines using the duplex of the mutant mtDNA assay and reference mtDNA assay shown in Fig. 3A. The percentage of mutant mtDNA was quantified by dividing the concentration (copies/μl) of high-amplitude FAM-positive droplets by the concentration of HEX-positive droplets. The percentage of WT mtDNA was quantified by dividing the concentration of low-amplitude FAM-positive droplets by the concentration of HEX-positive droplets. C, correlation analysis comparing the three standard methods for heteroplasmy quantification (Sanger sequencing, PspOMI ”Last cycle hot” PCR/RFLP, and HaeIII ”Last cycle hot” PCR/RFLP) to dPCR. The R value comparing dPCR to Sanger sequencing was 0.9938, dPCR to PspOMI ”Last cycle hot” PCR/RFLP was 0.9924, and dPCR to HaeIII ”Last cycle hot” PCR/RFLP was 0.9954. mtDNA, mitochondrial DNA; dPCR, digital PCR; RFLP, restriction fragment-length polymorphism.