Figure 6.
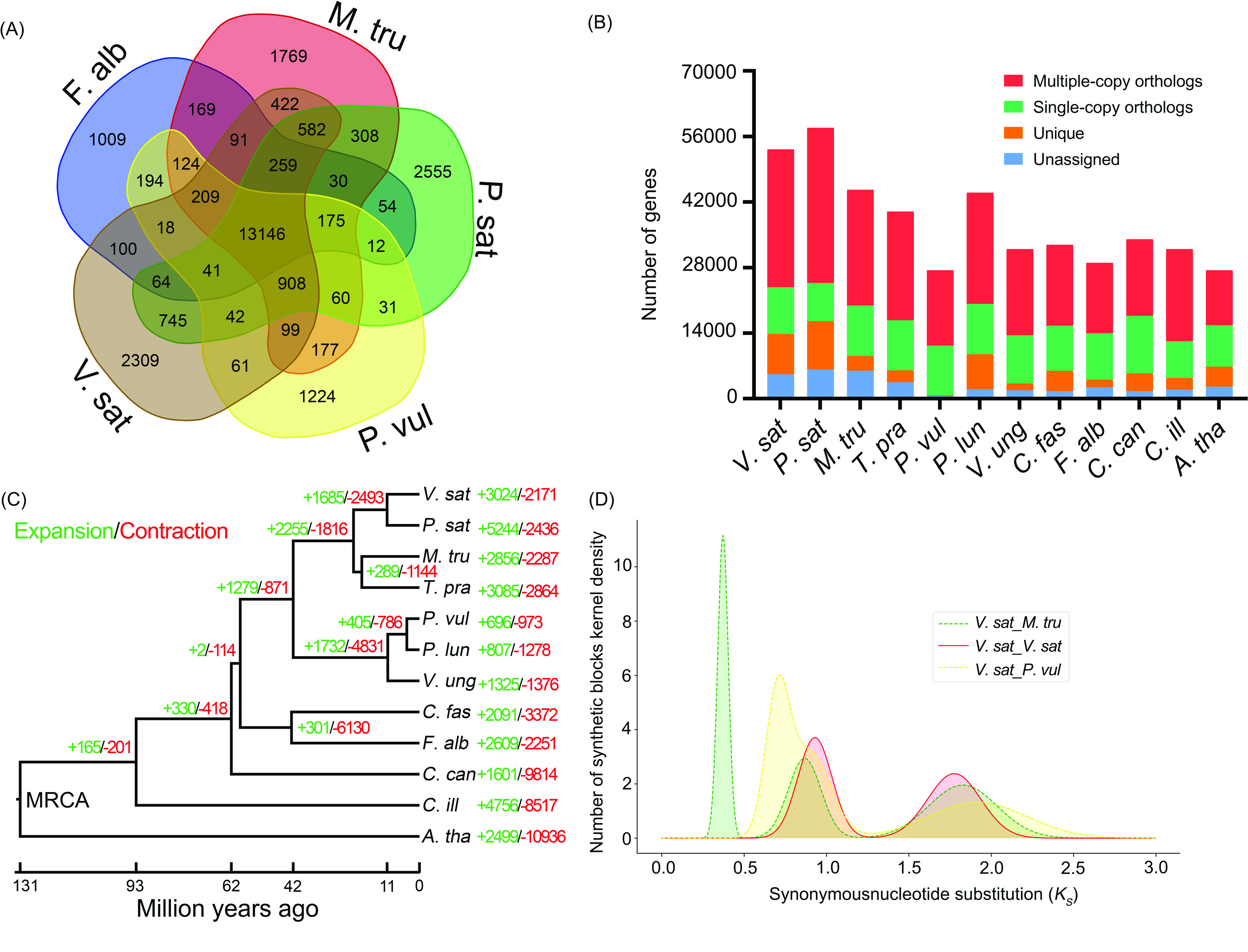
Evolution of the V. sativa genome. (A) A Venn diagram showing shared and unique orthologous gene families in V. sativa and four other legumes. (B) Predicted orthologous protein composition for V. sativa compared to A. thaliana, C. illinoinensis and nine legumes. (C) A phylogenetic tree shows the expansion and contraction of the gene families and the divergence time for species. (D) Ks plot shows the whole genome duplication event in V. sativa, M. truncatula and P. vulgaris. V. sat: Vicia sativa, P. sat: Pisum sativum, M. tru: Medicago truncatula, T. pra: Trifolium pratense, P. vul: Phaseolus vulgaris, P. lun: Phaseolus lunatus, V. ung: Vigna unguiculata, C. fas: Chamaecrista fasciculata, F. alb: Faidherbia albida, C. can: Cercis canadensis, C. ill: Carya illinoinensis, A. tha: Arabidopsis thaliana.
