Figure 1. Approaches to genetic screening.
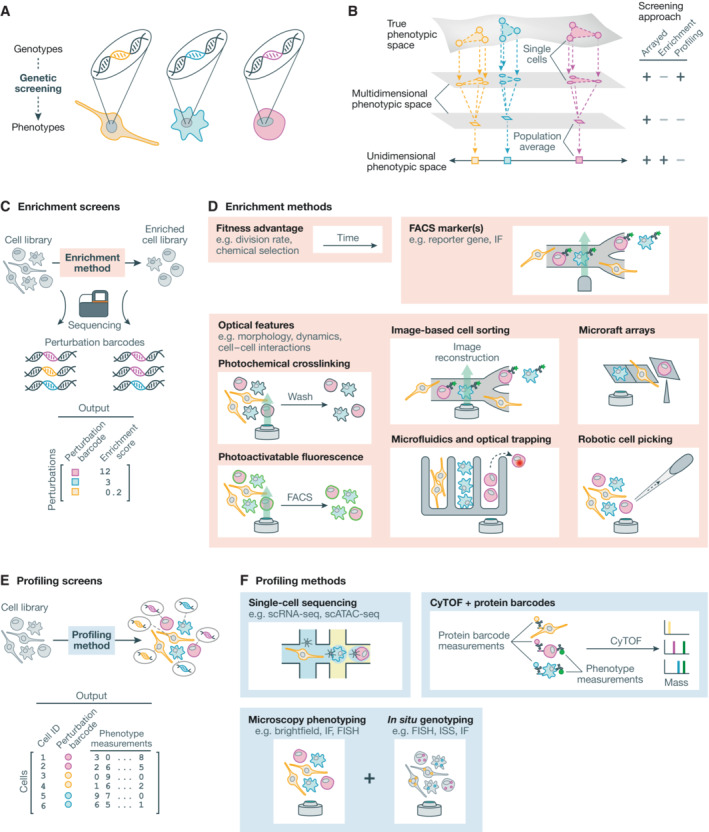
(A) Genetic screens seek to map genotypes to the phenotypes they produce. (B) Screening methodologies capture projections of cell phenotypes. Pooled profiling screens project individual cells into a multidimensional phenotypic space defined by the profiling method. Pooled enrichment screens project population averages into a unidimensional phenotypic space defined by the enrichment criteria. Arrayed screens can embody either of these phenotype–genotype associations. (C) Enrichment screens subject an initial cell library to an enrichment process to select for a phenotype of interest. Perturbation enrichment is determined by comparing the abundance of perturbation barcodes in the cell library before and after selection using next generation sequencing. (D) Cells can be enriched through a fitness advantage, fluorescence‐activated cell sorting, or one of several approaches to isolate cells based on microscopy‐defined features. (E) Profiling screens subject a complete cell library to profiling. Individual cells are assigned both perturbations and multidimensional phenotypic measurements. (F) Single cell profiling methods for genetic screening include single‐cell sequencing approaches, CyTOF using protein barcodes, and microscopy‐based phenotyping with in situ genotyping. FACS, fluorescence‐activated cell sorting; IF, immunofluorescence; scRNA‐seq, single‐cell RNA sequencing; scATAC‐seq, single‐cell assay for transposase‐accessible chromatin using sequencing; CyTOF, cytometry by time‐of‐flight.
