Figure 4. Optical methods for multidimensional phenotypic profiling.
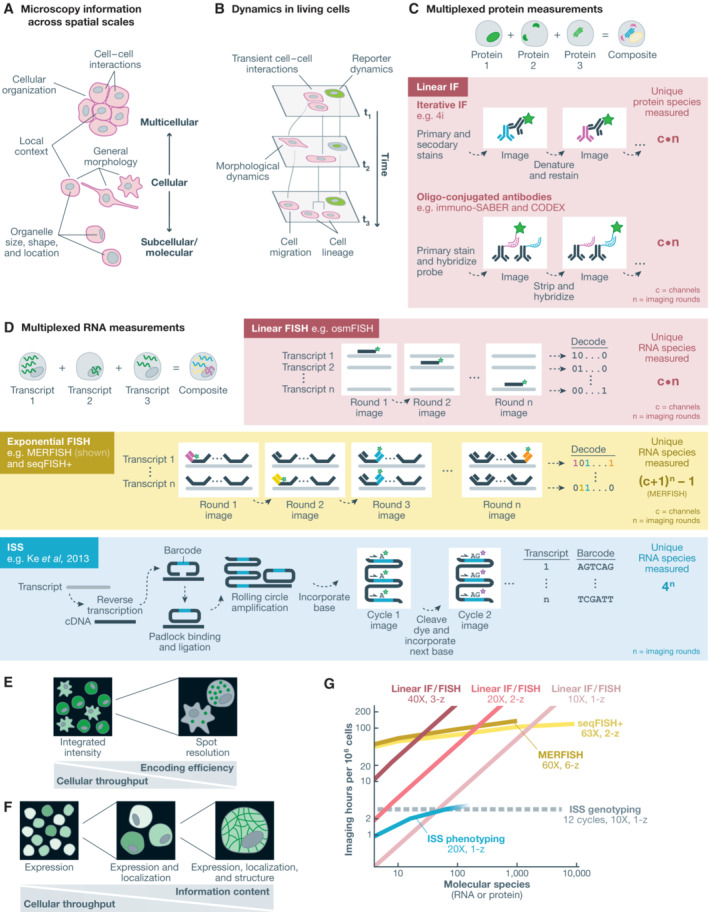
(A) Microscopy can capture molecular, subcellular, cellular, and multicellular phenotypes. (B) Live cell microscopy captures cellular dynamics. (C) Multiplexed protein measurements enable the observation of multiple proteins in each cell. Iterative immunofluorescence approaches rely on multiple rounds of sample staining with dye‐conjugated antibodies and fluorescence imaging. Oligo‐conjugated antibodies enable measurement with hybridization of fluorescence in situ hybridization probes. (D) Multiplexed RNA measurements enable the observation of multiple RNA species in each cell through either fluorescence in situ hybridization (FISH) or in situ sequencing (ISS). Linear FISH approaches encode RNA measurements with a linear encoding relative to imaging iterations. Exponential FISH techniques facilitate measurement of an exponentially increasing number of RNA species with a linear increase in imaging iterations. ISS approaches similarly enable exponential encoding of RNA species across sequencing cycles. Encoding efficiencies are theoretical and do not account for additional imaging cycles/rounds used for error correction in exponential‐scaling techniques. (E) RNA measurements requiring spot resolution can efficiently encode many RNA species but require high‐magnification imaging, while linear encoding methods can rely on integrated intensity measurements, instead of spot resolution, to enable high throughput at lower optical magnification. (F) Spatial detail and imaging time both increase as protein localization is imaged at higher magnification, presenting a tradeoff between cellular throughput and information content. (G) Estimated imaging throughputs and multiplexing capacities for selected multiplexed RNA and protein measurement approaches, based on magnification and z‐stack requirements, fluorescence channels, imaging rounds, and theoretical encoding efficiency without error correction. For comparison, the required imaging time to genotyping one million cells with 12 cycles of in situ sequencing is shown in gray. At high multiplexes, ISS phenotyping becomes less quantitative due to optical crowding. We do not consider expansion microscopy approaches here, which can increase the maximum multiplex for resolution‐limited techniques at the cost of increased imaging time. IF, immunofluorescence; FISH, fluorescence in situ hybridization; ISS, in situ sequencing.
