Abstract
Background
Genogroup II noroviruses are the most common cause of acute infectious gastroenteritis. We evaluated the use of a new GII.2 inoculum in a human challenge.
Methods
Forty-four healthy adults (36 secretor-positive and 8 secretor-negative for histo-blood group antigens) were challenged with ascending doses of a new safety-tested Snow Mountain virus (SMV) GII.2 norovirus inoculum (1.2 × 104 to 1.2 × 107 genome equivalent copies [GEC]; n = 38) or placebo (n = 6). Illness was defined as diarrhea and/or vomiting postchallenge in subjects with evidence of infection (defined as GII.2 norovirus RNA detection in stool and/or anti-SMV immunoglobulin G [IgG] seroconversion).
Results
The highest dose was associated with SMV infection in 90%, and illness in 70% of subjects with 10 of 12 secretor-positive (83%) and 4 of 8 secretor-negative (50%) becoming ill. There was no association between prechallenge anti-SMV serum IgG concentration, carbohydrate-binding blockade antibody, or salivary immunoglobulin A and infection. The median infectious dose (ID50) was 5.1 × 105 GEC.
Conclusions
High rates of infection and illness were observed in both secretor-positive and secretor-negative subjects in this challenge study. However, a high dose will be required to achieve the target of 75% illness to make this an efficient model for evaluating potential norovirus vaccines and therapeutics.
Clinical Trials Registration
Keywords: norovirus, human challenge, Snow Mountain virus, viral gastroenteritis, infectious dose, ID50
We evaluated the use of a new GII.2 inoculum in a human challenge that could facilitate the evaluation of norovirus vaccines and therapeutics and observed high rates of infection and illness in both secretor-positive and -negative subjects.
Worldwide, noroviruses are the most common cause of endemic and epidemic acute gastroenteritis across all ages [1]. In the United States, norovirus causes up to 21 million illnesses, with 1.9 million outpatient visits and 71 000 hospitalizations annually [2]. Human noroviruses are subdivided into at least 10 genogroups with multiple genotypes and subgroups [3]. The most common genogroup associated with human infection is genogroup II (GII), with the Snow Mountain virus (SMV) being a prototype genotype GII.2 strain.
Noroviruses are highly infectious, and the infectious dose required for transmission is low (<1500 viral particles or the equivalent of a median infectious dose [ID50] between 2.9 and 95 genome equivalent copies [GEC] in secretor-positive subjects in human challenge studies) [4, 5]. Transmission is via the fecal-oral route or via airborne droplets of vomitus containing norovirus [6]. Prior studies have shown that the risk of norovirus infection is oftentimes associated with the presence of a FUT-2 allele [7–9]. The FUT-2 allele is found in approximately 80% of people of northern European descent and encodes an α-1,2-fucosyltransferase responsible for the expression of the H type 1 histo-blood group antigens (HBGAs) on mucosal surfaces, in saliva, and in other body fluids (eg, secretor-positive) [10]. The HBGAs on the surfaces of the intestinal epithelial cells are postulated to facilitate the attachment and cell entry for some norovirus strains. Therefore, the absence of HGBA (ie, secretor-negative) is considered to be a strong protective factor against certain norovirus infections [11, 12].
Norovirus infection is characterized by a brief incubation period (24–48 hours) as well as a short duration of illness (12–60 hours) [13] in immunocompetent individuals. While asymptomatic norovirus infection can occur, symptomatic infection typically includes nausea, vomiting (nonbloody, nonbilious), watery diarrhea (nonbloody), abdominal pain, and systemic symptoms (generalized myalgias, malaise, headache, and fever). Severe manifestations occur mostly in immunocompromised patients and at the extremes of age.
Currently, there is no specific treatment or prevention for norovirus. Human challenge models for norovirus have been described for genogroup I.1 and genogroup II.1, II.2, and II.4 [4, 14–16]. Due to the importance of GII in norovirus outbreaks [17], it is critical to develop and characterize norovirus GII challenge inocula that can be used to evaluate potential vaccines and therapeutics and to gain a better understanding of GII infectivity and disease mechanisms. The goals of this study were to (1) examine the dose-response relationship for infection and illness due to SMV (GII.2), (2) study the immune responses after challenge, and (3) explore the role of secretor status on SMV infection and illness.
MATERIALS AND METHODS
Study Design
This is the first randomized, double blind, placebo-controlled study of a new human challenge stock of SMV (norovirus genogroup II, genotype 2 [GII.2]). Healthy males and nonpregnant females between the ages of 18 and 49 years were enrolled if they met all eligibility criteria (NCT02473224). Subjects were followed for safety, illness, infection, and immune responses with collection of blood, saliva, emesis, and stool. The study was conducted at Emory University and approved by the Emory Institutional Review Board. Written informed consent was obtained from all subjects before enrollment (Supplementary Materials).
Challenge and SMV Stock
The clinical grade product was obtained from Ralph Baric at the University of North Carolina at Chapel Hill (lot 001-09SM, Investigational New Drug [IND] 14697) (Supplementary Materials).
Randomization and Study Groups
The study consisted of 4 sequential cohorts of 11 subjects and used an adaptive design intended to achieve illness in about 75% of subjects with 1 of 3 oral doses of the challenge stock starting with 1.2 × 104 GEC. Four cohorts of secretor-positive subjects were enrolled for ascending dose challenge. The last cohort also included secretor-negative subjects to determine the role of secretor status on susceptibility to infection and illness. Therefore, we describe here the results from 5 groups: (1) low challenge dose with secretor-positive status (1.2 × 104 GEC, n = 9); (2) medium challenge dose with secretor-positive status (1.2 × 106 GEC, n = 9); (3) high challenge dose (1.2 × 107 GEC, n = 12) with secretor-positive status; (4) high challenge dose (1.2 × 107 GEC, n = 8) with secretor-negative status; and (5) placebo (n = 6). The study safety monitoring committee reviewed the safety data from the prior cohort and approved continuation of the study before the next cohort was enrolled.
Assays
Detection and quantitation of SMV RNA in stool was performed as previously described [18] (Supplementary Materials).
Secretor phenotyping was determined by testing saliva samples collected at screening for the presence of salivary H type 1 carbohydrate as previously described [19] (Supplementary Materials). Detection of anti-SMV immunoglobulin G (IgG) in serum and immunoglobulin A (IgA) in saliva were detected by quantitative enzyme-linked immunosorbent assays [8, 20] (Supplementary Materials). The SMV virus-like particle (VLP) carbohydrate-binding antibody blockade assay was developed based on previously reported Norwalk virus and norovirus GII.4 blockade assays [21, 22] (Supplementary Materials).
Definitions
Illness was defined as diarrhea (≥3 loose or liquid stools per 24 hours, ie, takes the shape of the container, or ≥300 g of loose or liquid stools per 24 hours), and/or vomiting during the inpatient period, in a subject with evidence of infection. Infection was defined as virus detection in stool by reverse-transcription quantitative polymerase chain reaction (RT-qPCR) with SMV-specific primers and probe as previously described [18] and/or seroconversion (defined as a 4-fold rise from prechallenge in SMV IgG titer in any postchallenge serum sample through day 30).
Statistical Analysis
Categorical variables are summarized with counts and percentages. The 95% confidence intervals (CIs) for these variables were constructed using exact Clopper–Pearson methods. Continuous variables are summarized with mean and standard deviation or median and interquartile range, where appropriate. For all assays, the geometric mean and 95% CIs (based on Student t distribution) are presented. The ID50 (human infectious dose causing 50% infection) was determined using a logistic regression model among secretor-positive subjects. Multivariable logistic regression models were fit with binary infection status as the outcome and the following covariates: sex, race, prechallenge antibody concentration, dose, secretor status, and age to estimate the effect of prechallenge titers on the probability of infection while controlling for potential confounders. A separate model was used for each type of antibody assay. All data analyses and presentations were conducted with SAS software, version 9.4 (SAS Institute, Cary, North Carolina).
RESULTS
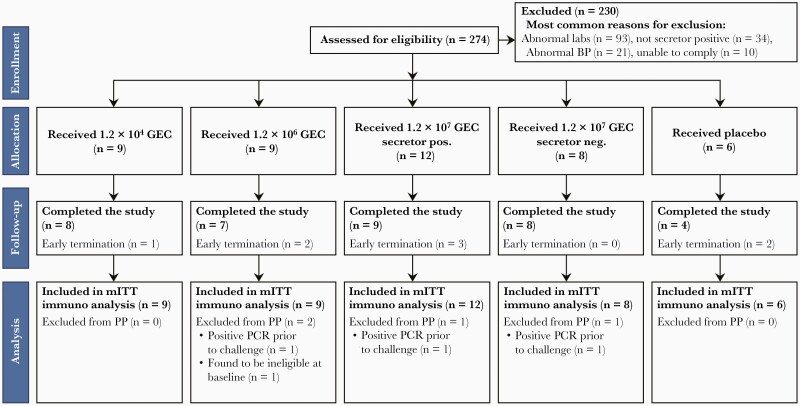
A total of 274 subjects were screened, and 44 were enrolled between 8 March 2016 and 13 June 2018. The 2 most common reasons for screen failure were the abnormal laboratory screening results (for all 4 cohorts) or saliva secretor status for the first 3 cohorts. Subjects were randomly assigned to receive SMV (n = 38) or placebo (n = 6). Thirty-six enrolled subjects successfully completed the entire study protocol (Figure 1). The mean age of the study population was 32.6 years (range, 20–49 years) and the majority were male (59%) with 61% Black/African American or multiracial and 9% Hispanic or Latino (Supplementary Table 1).
Figure 1.
Consolidated Standards of Reporting Trials (CONSORT) flow diagram. The following laboratory tests were performed at screening: white blood cells, hemoglobin, platelets, absolute neutrophil count, creatinine, alanine aminotransferase, total bilirubin, potassium, sodium, hemoglobin A1c, serologies for human immunodeficiency virus, hepatitis B, hepatitis C, urine protein, stool culture, norovirus stool by reverse-transcription quantitative polymerase chain reaction, and microscopic examination of the stool for ova and parasites. Abbreviations: BP, blood pressure; GEC, genome equivalent copies; mITT, modified intention-to-treat; PCR, polymerase chain reaction; PP, per protocol.
Clinical Presentation
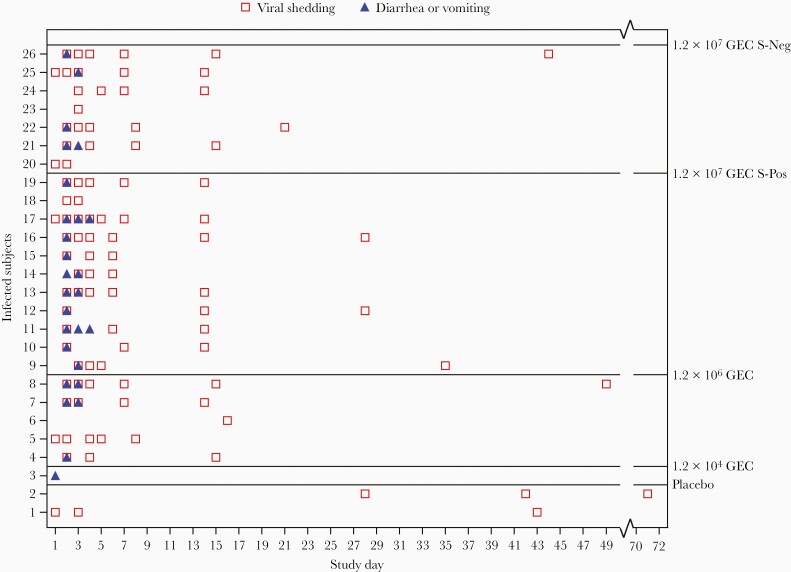
Overall, 24 of the 38 subjects (63%) who received the challenge stock became infected, and 75% of those who were infected were ill. At the high challenge dose, more subjects were ill (especially vomiting) if they were secretor-positive (83% [95% CI, 52%–98%) than secretor-negative (50% [95% CI, 16%–84%]), although infection rates for secretor-positive and secretor-negative subjects at the high dose were quite similar (92% vs 88%, respectively) (Table 1). In the medium challenge dose, 50% of secretor-positive subjects had either symptom (95% CI, 16%–84%) (Table 1). Vomiting/diarrhea were observed mainly on days 2–3 postchallenge (range, days 1–4) (Figure 2). Details on solicited adverse events are listed in Supplementary Table 2.
Table 1.
Proportion of Subjects With GII.2 Snow Mountain Virus–Associated Illness and/or Infection Following Challenge in the Modified Intention-to-Treat Population
| Status and Outcome | mITT Population | ||||
|---|---|---|---|---|---|
| 1.2 × 104 GECa (n = 9) | 1.2 × 106 GECa,b (n = 9) | 1.2 × 107 GEC S-Pos (n = 12) | 1.2 × 107 GEC S-Neg (n = 8) | Placebo (n = 6)c | |
| Gastrointestinal symptoms | |||||
| Diarrhead | 0 (0) | 1 (13) | 4 (33) | 2 (25) | 0 (0) |
| Vomiting | 1 (11) | 4 (50) | 10 (83) | 3 (38) | 0 (0) |
| Diarrhea and/or vomiting | 1 (11) | 4 (50) | 10 (83) | 4 (50) | 0 (0) |
| Infection | |||||
| Sheddinge | 0 (0) | 5 (56) | 11 (92) | 7 (88) | 2 (33) |
| Seroconversion | 1 (11) | 3 (33) | 7 (58) | 6 (75) | 1 (17) |
| Shedding and/or seroconversion | 1 (11) | 5 (56) | 11 (92) | 7 (88) | 2 (33) |
| Illness | |||||
| Shedding and/or seroconversion with diarrhea and/or vomiting | 1 (11) | 3 (38) | 10 (83) | 4 (50) | 0 (0) |
Data are presented as No. (%).
Abbreviations: GEC, genome equivalent copies; mITT, modified intention-to-treat population; S-Neg, secretor-negative; S-Pos, secretor-positive.
1.2 × 104 GEC and 1.2 × 106 GEC were only administered to S-Pos.
One subject in the 1.2 × 106 GEC group left the inpatient unit on day 2 without reporting any stool data or vomiting episodes; therefore, the denominator for these outcomes is 1 fewer than the mITT population.
Two subjects who received placebo met the definition of infection. The first subject had detectable virus in stool on days 1 and 3. The second subject had both detectable virus in stool and a 4-fold rise in Snow Mountain virus–specific serum immunoglobulin G on day 28.
Diarrhea defined as (≥3 loose or liquid stools (ie, takes the shape of the container), or ≥300 g of loose or liquid stools per 24 hours) during the inpatient period. The 24-hour period is defined as a study day from 12:00 Am to 11:59 Pm.
Shedding defined as excreting challenge virus in stool any time after challenge through day 30. Seroconversion defined as at least a 4-fold rise in SMV-specific serum IgG from baseline through day 30.
Figure 2.
Duration of RNA positivity and symptoms in subjects with GII.2 Snow Mountain virus infection following challenge in the modified intention-to-treat population. 1.2 × 104 genome equivalent copies (GEC) and 1.2 × 106 GEC were only administered to secretor-positive subjects. Abbreviations: GEC, genome equivalent copies; S-Neg, secretor-negative; S-Pos, secretor-positive.
Infection: SMV RNA Shedding
Number of subjects with viral shedding in stool was dose dependent; however, peak virus concentration in stool was similar for the medium- and high-dose challenge groups with 9.4 (95% CI, 5.09–13.65) and 9.8 (95% CI, 8.68–10.90) log10 GEC/g, respectively. Peak viral concentrations occurred a day earlier in the high-dose challenge group (Table 2).
Table 2.
Detection of Snow Mountain Virus by Reverse-Transcription Polymerase Chain Reaction in Stool, Modified Intention-to-Treat Population
| Parameter | Statistica | mITT Population | ||||
|---|---|---|---|---|---|---|
| 1.2 × 104 GEC (n = 9) | 1.2 × 106 GEC (n = 9) |
1.2 × 107 GEC S-Pos (n = 12) | 1.2 × 107 GEC S-Neg (n = 8) | Placebo (n = 6) | ||
| Cumulative virus shedding during inpatient period (log10 GEC) | n.; mean (SD) | 0 | 4; 11.4 (3.1) | 11; 11.6 (1.7) | 7; 11.4 (3.4) | … |
| Day of peak virus shedding per day during inpatient period | n.; median (range) | 0 | 4; 3.0 (2–4) | 11; 3.0 (2–4) | 7; 3.0 (2–5) | 1; 3.0 (3–3) |
| Peak virus concentration (log10 GEC/g)a | n.; mean (SD) | 1; 4.5 (NC) | 5; 9.4 (3.4) | 12; 9.8 (1.7) | 7; 9.8 (3.3) | 2; 7.2 (1.1) |
| Day of peak virus concentration | n.; median (range) | 1; 49.0 (49–49) | 5; 4.0 (3–16) | 12; 3.0 (2–48) | 7; 3.0 (1–8) | 2; 15.5 (3–28) |
| Duration of virus shedding (days) | n.; median (range) | 1; 1.0 (1–1) | 5; 13.0 (1–48) | 12; 13.0 (2–33) | 7; 14.0 (1–43) | 2; 43.5 (43–44) |
Abbreviations: GEC, genome equivalent copies; mITT, modified intention-to-treat population; NC, not calculable; S-Neg, secretor-negative; S-Pos, secretor-positive.
n value = restricted to subjects who were shedding on at least 1 day.
Interestingly, SMV RNA was detected in fecal specimens from 2 of 6 subjects in the placebo group. The first subject had detectable SMV RNA in stool on days 1, 3, and 45 postchallenge without seroconversion or a rise in carbohydrate blockade antibody titer and the second subject had SMV RNA in stool (days 28, 42, and 71 postchallenge) and a >4-fold rise in SMV-specific serum IgG on days 28 and 42 postchallenge, as well as a 32-fold rise in carbohydrate blockade antibody titer (Table 1, Figure 2).
IgG Seroconversion
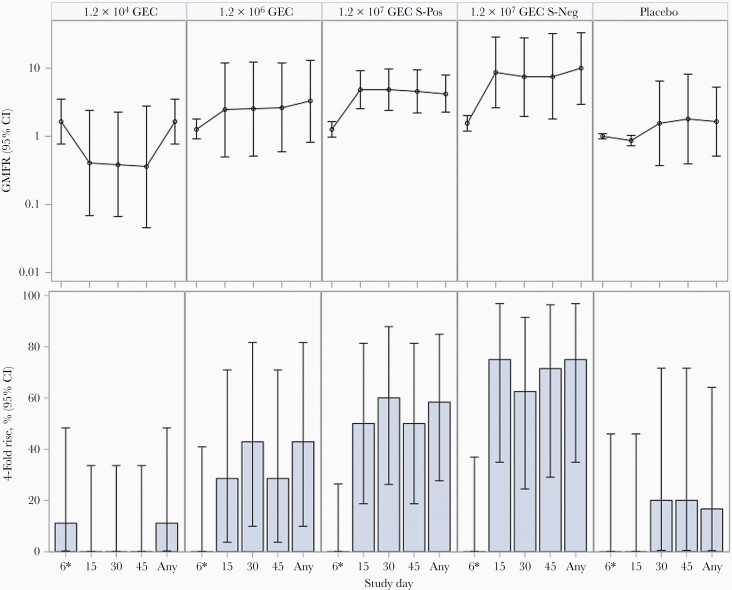
No difference was observed in frequency of seroconversion (>4-fold increase in anti-SMV IgG titers compared to prechallenge) when the secretor-positive subjects were compared to the secretor-negative subjects receiving the same dose or when comparing subjects who received either the high or medium challenge dose. Only 1 subject seroconverted by the time of discharge from the inpatient unit (day 5). Seroconversion occurred in the medium and high challenge dose groups starting around day 15 postchallenge (Supplementary Table 3, Figure 3).
Figure 3.
Serum Snow Mountain virus immunoglobulin G geometric mean fold rise and 4-fold rise by time point and treatment group. 1.2 × 104 genome equivalent copies (GEC) and 1.2 × 106 GEC were only administered to secretor-positive subjects. Abbreviations: CI, confidence interval; GEC, genome equivalent copies; GMFR, geometric mean fold rise; S-Neg, secretor-negative; S-Pos, secretor-positive.
Norovirus-Associated Illness
At the high challenge dose, illness (vomiting and/or diarrhea plus shedding and/or seroconversion) occurred in 10 of 12 (83% [95% CI, 52%–98%]) secretor-positive subjects compared to 4 of 8 (50% [95% CI, 16%–84%]) secretor-negative subjects. At the medium challenge dose, 3 of 9 secretor-positive subjects (33% [95% CI, 52%–100%]) had norovirus-associated illness, while 1 of 9 secretor-positive subjects (11% [95% CI, 0–48%]) had norovirus-associated illness in the low challenge dose group. No subjects in the placebo group had illness, including the 2 subjects who had SMV RNA detected in fecal specimens (95% CI, 0–46%) (Table 1).
Safety
The SMV challenge had an acceptable safety profile. None of the subjects in any group experienced a solicited symptom that met serious adverse events criteria or an unsolicited adverse event (AE) that was grade 3 after challenge through day 30. A total of 2 subjects experienced grade 3 clinical laboratory AEs. One secretor-negative subject experienced severe neutropenia (<900 cells/µL) on day 5 that was determined to be related to the study product and was asymptomatic and spontaneously resolved, and 1 placebo subject experienced severe hypokalemia on day 15; this subject did not have detectable SMV RNA in stool.
Immune Responses
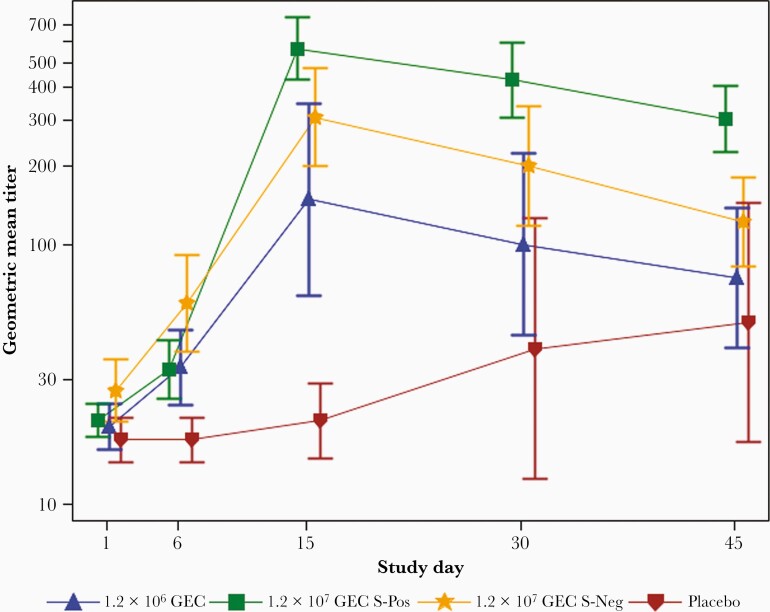
Serum antibodies that blocked binding of SMV VLPs to blood type B-PAA-biotin carbohydrate (carbohydrate blockade antibodies) were measured as a proxy for neutralizing antibodies, and a high percentage of seroconversion was observed in the medium (57%) and high (in 75% of secretor-negatives and 100% of secretor-positives) challenge dose groups by day 15. The geometric mean titer (GMT) for the SMV carbohydrate blockade antibody peaked at day 15 with the highest levels in the high challenge dose secretor-positive group (566 [95% CI, 302–1062]) (Figure 4) as compared to GMT of subjects from other groups. Subjects who received the low dose were not evaluated for carbohydrate blockade antibody response because of lack of evidence of infection.
Figure 4.
Geometric mean titer of Snow Mountain virus (SMV)–specific carbohydrate blockade serum antibody over time by treatment group. Cohort challenged with 1.2 × 104 genome equivalent copies (GEC) was not tested for SMV-specific carbohydrate blockade serum as no subject developed illness. Error bars represent standard error. Abbreviations: GEC, genome equivalent copies; S-Neg, secretor-negative; S-Pos, secretor-positive.
Rise in anti-SMV salivary IgA concentration relative to total salivary IgA concentration first occurred at day 15, but overall, salivary IgA conversion (defined as ≥4-fold rise in the ratio of SMV-specific salivary IgA to total salivary IgA at any timepoint compared to prechallenge saliva) was observed in fewer study subjects compared to serum IgG seroconversion. Both secretor-negative and secretor-positive subjects who received the high challenge dose had a similar percentage with salivary IgA conversion at any time point: 38% (95% CI, 9%–76%) and 42% (95% CI, 15%–72%), respectively. No subjects in the low challenge dose group had an increase in anti-SMV salivary IgA, and only 1 subject each in the medium challenge dose group and the placebo group demonstrated an SMV-specific salivary IgA conversion (Table 3).
Table 3.
Saliva Snow Mountain Virus Immunoglobulin A (IgA) Over Total IgA Geometric Mean Fold Rise and Seroresponse (4-Fold Rise) Results by Time Point and Treatment Group, Modified Intention-to-Treat Population
| Study Day | Statistic | SMV Challenge Dose (GEC), mITT Population | ||||
|---|---|---|---|---|---|---|
| 1.2 × 104 (n = 9) | 1.2 × 106 (n = 9) | 1.2 × 107 S-Pos (n = 12) |
1.2 × 107 S-Neg (n = 8) |
Placebo (n = 6) | ||
| 2 | n; GMFRa (95% CI) | 9; 0.8 (.5–1.2) | 9; 1.0 (.8–1.3) | 12; 1.0 (.9–1.2) | 8; 1.2 (.7–1.8) | 6; 1.1 (.5–2.2) |
| 4-fold riseb (95% CI) | 0 (0–34) | 0 (0–34) | 0 (0–26) | 0 (0–37) | 0 (0–46) | |
| 3 | n; GMFRa (95% CI) | 9; 0.8 (.6–1.0) | 8; 1.0 (.8–1.3) | 12; 1.3 (1.0–1.6) | 8; 0.9 (.7–1.3) | 6; 0.6 (.3–1.2) |
| 4-fold riseb (95% CI) | 0 (0–34) | 0 (0–37) | 0 (0–26) | 0 (0–37) | 0 (0–46) | |
| 4 | n; GMFRa (95% CI) | 9; 0.8 (.5–1.2) | 7; 0.9 (.6–1.2) | 12; 0.9 (.7–1.2) | 8; 1.0 (.7–1.4) | 6; 1.0 (.6–1.8) |
| 4-fold riseb (95% CI) | 0 (0–34) | 0 (0–41) | 0 (0–26) | 0 (0–37) | 0 (0–46) | |
| 5 | n; GMFRa (95% CI) | 9; 0.9 (.6–1.3) | 7; 0.7 (.5–1.0) | 12; 1.0 (.7–1.2) | 8; 0.8 (.5–1.3) | 6; 0.9 (.4–2.0) |
| 4-fold riseb (95% CI) | 0 (0–34) | 0 (0–41) | 0 (0–26) | 0 (0–37) | 0 (0–46) | |
| 6 | n; GMFRa (95% CI) | 9; 0.9 (.6–1.3) | 7; 0.9 (.7–1.2) | 12; 1.2 (.8–1.7) | 8; 1.2 (.8–1.8) | 6; 1.0 (.6–1.8) |
| 4-fold riseb (95% CI) | 0 (0–34) | 0 (0–41) | 0 (0–26) | 0 (0–37) | 0 (0–46) | |
| 15 | n; GMFRa (95% CI) | 9; 0.6 (.4–.9) | 7; 1.7 (.8–3.7) | 12; 4.2 (2.3–7.9) | 8; 3.0 (1.4–6.3) | 6; 1.1 (.7–1.8) |
| 4-fold riseb (95% CI) | 0 (0–34) | 14 (0–58) | 42 (15–72) | 38 (9–76) | 0 (0–46) | |
| 30 | n; GMFRa (95% CI) | 9; 0.5 (.3–.9) | 7; 1.3 (.7–2.3) | 12; 2.7 (1.6–4.8) | 8; 2.1 (1.1–3.8) | 5; 1.4 (.6–3.4) |
| 4-fold riseb (95% CI) | 0 (0–34) | 14 (0–58) | 33 (10–65) | 25 (3–65) | 20 (1–72) | |
| 45 | n; GMFRa (95% CI) | 9; 0.8 (.5–1.2) | 7; 2.1 (1.2–3.8) | 10; 2.2 (1.2–4.2) | 7; 1.4 (.7–2.8) | 5; 1.3 (.6–2.8) |
| 4-fold riseb (95% CI) | 0 (0–34) | 14 (0–58) | 20 (3–56) | 14 (0–58) | 0 (0–52) | |
| Any time | n; GMFRa (95% CI) | 9; 1.3 (.9–1.7) | 9; 2.3 (1.5–3.5) | 12; 4.4 (2.4–8.0) | 8; 3.4 (1.9–6.2) | 6; 1.8 (1.0–3.2) |
| 4-fold riseb (95% CI) | 0 (0–34) | 11 (0–48) | 42 (15–72) | 38 (9–76) | 17 (0–64) | |
Abbreviations: CI, confidence interval; GEC, genome equivalent copies; GMFR, geometric mean fold rise; mITT, modified intention-to-treat; SMV, Snow Mountain virus; S-Neg, secretor-negative; S-Pos, secretor-positive.
GMFR in the ratio of SMV-specific immunoglobulin A (IgA) over total IgA compared to prechallenge (day 1). Any time represents the geometric mean of the maximum fold rise.
Four-fold rise represents the percentage of subjects with at least a 4-fold rise in the ratio of SMV-specific IgA over total IgA compared to prechallenge (day 1).
There was no clear association between infection status and prechallenge antibody concentrations for serum IgG, serum blockade antibody, or salivary IgA.
We calculated the ID50 (defined as the dose at which 50% of secretor-positive subjects are infected) to be 5.1 × 105 GEC (95% CI, 2.8 × 104 to 2.7 × 106) using a logistic regression model.
DISCUSSION
Noroviruses are characterized by high infectivity and environmental persistence and are responsible for substantial morbidity and societal and economic burdens. Live challenge model approaches in humans have been used in clinical development of biologics against enteric bacterial pathogens including enterotoxigenic Escherichia coli, Campylobacter jejuni, Vibrio cholerae, and Shigella spp. Few GII.2 norovirus challenge studies have been performed to date, and our study established the ID50 for this second-generation challenge stock as well as characterized infectivity and illness in secretor-negative individuals [4, 23, 24]. The overall infection (56%–92%) and illness (38%–83%) rates observed at the medium and high doses are consistent with previous reports for 2 GII.2 challenge trials with the first-generation SMV inoculum where infection rates ranged from 60% to 75% and illness rates ranged from 47% to 75% for a mixed population of secretor-positive and -negative subjects [4, 24]. Duration and magnitude of shedding and seroconversion rates were similar between the medium and high challenge doses and between secretor-positive or -negative subjects.
Susceptibility to norovirus is modulated in part by expression of specific, genetically determined carbohydrate HBGAs determining secretor status [8]. An earlier GI.1 norovirus virus challenge study reported that infection is secretor-dependent with 62% of the secretor-positive subjects infected following challenge vs no secretor-negative subjects [8]. GII.4 challenge studies reported different infection rates in secretor-positive and secretor-negative subjects (70% and 6%, respectively) [15]. In the only prior SMV challenge study where secretor status was reported, 66% of secretor-positive subjects and 33% (1 of 3) of secretor-negative subjects were infected and developed illness [23]. The infected and ill secretor-negative subject received 3.17 × 105 GEC whereas the other 2 secretor-negative subjects who did not get infected received much lower doses. Our study results are unique in that the majority of secretor-negative subjects shed virus (88%) or seroconverted (75%), and 50% developed illness at the highest challenge dose. A recent meta-analysis of norovirus infectivity showed that the estimated mean infection rate was 5 times higher in secretor-positive compared to secretor-negative for the GII viruses [4]. Whether decreased susceptibility to norovirus infection in our secretor-negative subjects would have been more obvious at the medium challenge dose is unknown as secretor-negative subjects were only included in the group receiving the highest challenge cohort. Though novel for a challenge study, this finding is not unexpected as there are reports of GII norovirus outbreaks that included cases among both secretor-positive and secretor-negative individuals [25, 26] that could be explained in part by GII.2 noroviruses binding to alternative receptors. Another factor that could affect susceptibility to norovirus challenge is prior immunity. Though previous studies have suggested that preexisting antibodies may be associated with immunity to norovirus infection [27], in our study we did not see an association between prechallenge concentrations of anti-SMV serum IgG, serum blockade antibody, or anti-SMV salivary IgA and infection status in part because SMV is not the dominant circulating strain, and it is unclear how long norovirus immunity lasts [28]. Protective factors against norovirus infection may not be due to serological response but to other components of the immune system (ie, innate and cellular adaptive immunity), HLA restrictions, or lack of receptors to the virus [23]. Although there is substantial evidence to suggest that serum antibodies have the ability to block the binding of norovirus VLPs to HBGAs [21, 22], almost all of the subjects in our study had low titers of blockade antibody in their prechallenge sera, so it was not possible to evaluate the protective effect of these antibodies against infection and illness.
In our study, symptoms associated with norovirus infection were consistent with the reports from previous challenge trials in which infected subjects experienced vomiting more frequently than diarrhea, accompanied by a constellation of symptoms that occurred soon after SMV challenge and resolved rapidly [18]. However, in our study and among secretor-positive subjects in the medium- and high-dose groups, viral shedding in the stool (median, 13–14 days depending on the dose) was longer than the median shedding previously described in a GII.4 challenge study [18] (mostly limited to the first week postchallenge) but shorter than a prior GI.1 challenge study [14] (median, 28 days) although no stool samples were collected in our study between days 15 and 30. Data on norovirus shedding generated by challenge studies facilitate estimates of transmission risk and provide evidence for the development of appropriate control measures, such as disinfection. Previous studies have reported a wide range of norovirus titers in stools. We measured a high magnitude of SMV RNA titer with an average of 9 log10 SMV GEC/g of stool after the medium or high challenge doses. These high shedding titers and prolonged duration of shedding have been proposed as drivers of the high rates of GII outbreaks [29].
The infectivity of this new GII.2 challenge stock was surprisingly low. The estimated ID50 of 5.1 × 105 GEC for this second-generation inoculum is about 100-fold higher than that estimated by Teunis et al [4] for the 2 previous SMV challenge trials with the original SMV inoculum prepared by Dolin et al [24]. Both inocula were titrated multiple times by the same laboratory using the same primers to confirm the actual stock titer, and titers were confirmed in vial remainders after challenging subjects. It is possible that this discrepancy is due to the different statistical approaches used to estimate the ID50, though recalculating the ID50 for our study using the same β Poisson methodology used for earlier studies [4] yielded similar results (2.74 × 105 GEC). Another likely possibility is that the inoculum preparation process led to a loss in infectivity. A difference between the effect of norovirus inocula was also observed in a second-generation Norwalk virus inoculum where the infectivity of the 2 inocula was similar, but subjects who received the new challenge stock (8FIIb) had significantly less severe symptoms than subjects who received the original 8FIIa Norwalk virus inoculum [30]. These observations have significant implications for the use of second-generation inocula in future norovirus vaccine efficacy trials and need to be considered when planning dose, sample size, and outcome measures. Future studies should examine and compare virus sequences in these challenge inocula.
Two of the 6 subjects who were challenged with a placebo met the study definition of infection, but neither had illness. One of these subjects had 2 norovirus-positive stool samples in the first 3 days postchallenge and another positive stool at day 45 postchallenge but did not seroconvert or develop symptoms. The other subject had norovirus-positive stool samples at days 28, 42, and 71 postchallenge and seroconverted at day 28 after administration of placebo. The timing of the shedding and seroconversion suggest that the second subject had a community-acquired norovirus. We were not able to confirm that either of these subjects was shedding the challenge virus by sequencing the virus in the stool samples as attempts to reamplify and sequence the RNA from the day 45 postchallenge stool, and several other stool samples were not successful.
Other limitations of our study include the enrollment of subjects in good health and in a specific age range. Though typical for controlled human challenge studies, these restrictions did not allow for examination of infection and disease in more vulnerable populations. We did not study the effect of ABO blood types on the susceptibility to the challenge stock. Though GII.4 norovirus challenge did not reveal any relationship between norovirus infection and ABO blood type [15], previous GI challenge trials have reported differences in infection by blood type [8, 31], and further information about this relationship for GII.2 norovirus would be valuable. Even though vomiting is the signature symptom of norovirus, we only measured viral shedding in the stool. A prior study confirmed the presence of virus in emesis correlating with the shedding in the stool after GII.2 challenge [32]. To study viral shedding and the titer of the inoculum, our study relied on quantitative detection of SMV genome copies by RT-qPCR, consistent with methods used in all other recent challenge studies. However, recently developed norovirus culture methods could yield better information by enumerating infectious particles in the inocula and estimate the infectious fraction of the RT-qPCR–titrated numbers of genome copies [12, 33]. The role of innate and adaptive cellular immunity in susceptibility to GII.2 infection and illness in this study is still under investigation.
This SMV challenge model will allow testing of current GI.1/GII.4 candidate vaccines to evaluate the cross-neutralizing and type of specific blockade antibodies needed for protection as well as the fundamental consequences of GII.2 pathogenesis in humans.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We acknowledge our participants and the following Hope Clinic and Vaccine and Treatment Evaluation Unit (VTEU) staff: Eileen Osinski, Brittany Robinson, April Zion, Mary Bower, Rijalda Deovic, Ariel Kay, Divyanshu Raheja, Regina Mosley, Shantricia Johnson-Colbert, Dori Purcel, Briyana Domjahn, Emily Presmanes, Eric Johnson, Ghina Alaaeddine, Sharon Curate-Ingram, Sonia Krengel, Tiffany Stivers, Charles Bailey, Rotrease Regan, Hollie Macenczak, Melinda Ogilvie, Dean Kleinhenz, Hannah Huston, Sirajud-Deen Talib, Joanne Sadowski, Dilshad Rafi Ahmed, Sara Jo Johnson, Ellen DeStefano, T. Jean Winter, Dawn Battle, Susan Rogers, Akinlolu Fasanmi, Adeyomi Bello, Christopher Huerta, Dong Li Wang, Yongxian Xu, Pam Lankford-Turner, Tamera Franks, Juliet Morales, Nayoka Rimann Jingru Zhao, Yerun Zhu, Justin Colwell, Hady Samaha, Mary Hart, Michele McCullough, Srilatha Edupuganti, Colleen F. Kelley, Alexis Ahonen, Amy C. Sherman, Jianguo Xu, and Cecilia Losada. We would also like to thank our independent safety monitors Bruce Ribner and David Rimland and the Georgia Clinical and Translational Science Alliance nurses and staff Barry Clark, Nadine Hines, Lucy Kugbila, Monica Lalor, Sandra Leonard, Shawanda Carter, Carmile Jerome, Debora Clem, and Jennifer Shamloo. Special thanks to members of the Moe Lab: Milagros Aldeco, Marisa Gallegos, and Yvonne Kienast. We would also like to thank members of the statistical and data management team Holly Baughman, Andy Hoy, Tejal Tailor, Patricia Abduragimova, and Cassandra Ballou. And from Division of Microbiology and Infectious Diseases (DMID), a special thanks to Suzanne Murray and Mohammed ElSafy, and Gabriele Feolo. We would also like to thank Dr Peter Teunis for his statistical advice.
Disclaimer. The opinions expressed in this article are those of the authors and do not reflect the view of the National Institute of Allergy and Infectious Diseases (NIAID), the National Institutes of Health (NIH), the Department of Health and Human Services, or the United States government.
Financial support. This work was supported by the National Center for Advancing Translational Sciences of the NIH (award number UL1TR002378); the NIAID (training grant numbers T32AI074492 and 5R38AI140299-02, and grant number R01 AI148260 to R. S. B.); Wellcome Trust (203268/Z/16/Z to R. S. B.); and the Division of Microbiology and Infectious Diseases to the Emory VTEU (HHSN272200800005C HHSN272201300018I, HHSN27200003, and HHSN27200018).
Contributor Information
Nadine Rouphael, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Allison Beck, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Amy E Kirby, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Pengbo Liu, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Muktha S Natrajan, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Lilin Lai, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Varun Phadke, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Juton Winston, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Vanessa Raabe, New York University Grossman School of Medicine and New York University Vaccine Center, New York, New York, USA.
Matthew H Collins, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Tigisty Girmay, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Alicarmen Alvarez, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Nour Beydoun, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Vinit Karmali, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Joanne Altieri-Rivera, Hope Clinic of the Emory Vaccine Center, Division of Infectious Diseases, Department of Medicine, School of Medicine, Emory University, Atlanta, Georgia, USA.
Lisa C Lindesmith, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Evan J Anderson, Division of Infectious Diseases, Department of Pediatrics, School of Medicine, Emory University, Atlanta, Georgia, USA.
Yuke Wang, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Jill El-Khorazaty, The Emmes Company, LLC, Rockville, Maryland, USA.
Carey Petrie, The Emmes Company, LLC, Rockville, Maryland, USA.
Ralph S Baric, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA.
Shahida Baqar, Division of Microbiology and Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Christine L Moe, Rollins School of Public Health, Emory University, Atlanta, Georgia, USA.
Mark J Mulligan, New York University Grossman School of Medicine and New York University Vaccine Center, New York, New York, USA.
References
- 1. Ahmed SM, Hall AJ, Robinson AE, et al. . Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14:725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hall AJ, Lopman BA, Payne DC, et al. . Norovirus disease in the United States. Emerg Infect Dis 2013; 19:1198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vinje J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 2015; 53:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Teunis PFM, Le Guyader FS, Liu P, Ollivier J, Moe CL.. Noroviruses are highly infectious but there is strong variation in host susceptibility and virus pathogenicity. Epidemics 2020; 32:100401. [DOI] [PubMed] [Google Scholar]
- 5. Teunis PFM, Moe CL, Liu P, et al. . Norwalk virus: how infectious is it? J Med Virol 2008; 80:1468–76. [DOI] [PubMed] [Google Scholar]
- 6. Barclay L, Park GW, Vega E, et al. . Infection control for norovirus. Clin Microbiol Infect 2014; 20:731–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hutson AM, Airaud F, LePendu J, Estes MK, Atmar RL.. Norwalk virus infection associates with secretor status genotyped from sera. J Med Virol 2005; 77:116–20. [DOI] [PubMed] [Google Scholar]
- 8. Lindesmith L, Moe C, Marionneau S, et al. . Human susceptibility and resistance to Norwalk virus infection. Nat Med 2003; 9:548–53. [DOI] [PubMed] [Google Scholar]
- 9. Nordgren J, Svensson L.. Genetic susceptibility to human norovirus infection: an update. Viruses 2019; 11:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Le Pendu J. Histo-blood group antigen and human milk oligosaccharides: genetic polymorphism and risk of infectious diseases. Adv Exp Med Biol 2004; 554:135–43. [DOI] [PubMed] [Google Scholar]
- 11. Haga K, Ettayebi K, Tenge VR, et al. . Genetic manipulation of human intestinal enteroids demonstrates the necessity of a functional fucosyltransferase 2 gene for secretor-dependent human norovirus infection. mBio 2020; 11:e00251-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ettayebi K, Tenge VR, Cortes-Penfield NW, et al. . New insights and enhanced human norovirus cultivation in human intestinal enteroids. mSphere 2021; 6:e01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Banyai K, Estes MK, Martella V, Parashar UD.. Viral gastroenteritis. Lancet 2018; 392:175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Atmar RL, Opekun AR, Gilger MA, et al. . Norwalk virus shedding after experimental human infection. Emerg Infect Dis 2008; 14:1553–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frenck R, Bernstein DI, Xia M, et al. . Predicting susceptibility to norovirus GII.4 by use of a challenge model involving humans. J Infect Dis 2012; 206:1386–93. [DOI] [PubMed] [Google Scholar]
- 16. Atmar RL, Bernstein DI, Harro CD, et al. . Norovirus vaccine against experimental human Norwalk virus illness. N Engl J Med 2011; 365:2178–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Centers for Disease Control and Prevention. Norovirus activity—United States, 2006-2007. MMWR Morb Mortal Wkly Rep 2007; 56:842–6. [PubMed] [Google Scholar]
- 18. Kirby AE, Shi J, Montes J, Lichtenstein M, Moe CL.. Disease course and viral shedding in experimental Norwalk virus and Snow Mountain virus infection. J Med Virol 2014; 86:2055–64. [DOI] [PubMed] [Google Scholar]
- 19. Leon JS, Kingsley DH, Montes JS, et al. . Randomized, double-blinded clinical trial for human norovirus inactivation in oysters by high hydrostatic pressure processing. Appl Environ Microbiol 2011; 77:5476–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Monroe SS, Stine SE, Jiang X, Estes MK, Glass RI.. Detection of antibody to recombinant Norwalk virus antigen in specimens from outbreaks of gastroenteritis. J Clin Microbiol 1993; 31:2866–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reeck A, Kavanagh O, Estes MK, et al. . Serological correlate of protection against norovirus-induced gastroenteritis. J Infect Dis 2010; 202:1212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Atmar RL, Bernstein DI, Lyon GM, et al. . Serological correlates of protection against a GII.4 norovirus. Clin Vaccine Immunol 2015; 22:923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindesmith L, Moe C, Lependu J, Frelinger JA, Treanor J, Baric RS.. Cellular and humoral immunity following Snow Mountain virus challenge. J Virol 2005; 79:2900–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dolin R, Reichman RC, Roessner KD, et al. . Detection by immune electron microscopy of the Snow Mountain agent of acute viral gastroenteritis. J Infect Dis 1982; 146:184–9. [DOI] [PubMed] [Google Scholar]
- 25. Lindesmith LC, Brewer-Jensen PD, Mallory ML, et al. . Virus-host interactions between nonsecretors and human norovirus. Cell Mol Gastroenterol Hepatol 2020; 10:245–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mallory ML, Lindesmith LC, Brewer-Jensen PD, Graham RL, Baric RS.. Bile Facilitates human norovirus interactions with diverse histoblood group antigens, compensating for capsid microvariation observed in 2016-2017 GII.2 strains. Viruses 2020; 12:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wyatt RG, Dolin R, Blacklow NR, et al. . Comparison of three agents of acute infectious nonbacterial gastroenteritis by cross-challenge in volunteers. J Infect Dis 1974; 129:709–14. [DOI] [PubMed] [Google Scholar]
- 28. Parra GI, Squires RB, Karangwa CK, et al. . Static and evolving norovirus genotypes: implications for epidemiology and immunity. PLoS Pathog 2017; 13:e1006136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matthews JE, Dickey BW, Miller RD, et al. . The epidemiology of published norovirus outbreaks: a review of risk factors associated with attack rate and genogroup. Epidemiol Infect 2012; 140:1161–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu P, Rahman M, Leon J, Moe C.. Less severe clinical symptoms of Norwalk virus 8fIIb inoculum compared to its precursor 8fIIa from human challenge studies. J Med Virol 2021; 93:3557–63. [DOI] [PubMed] [Google Scholar]
- 31. Hutson AM, Atmar RL, Graham DY, Estes MK.. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis 2002; 185:1335–7. [DOI] [PubMed] [Google Scholar]
- 32. Kirby AE, Streby A, Moe CL.. Vomiting as a symptom and transmission risk in norovirus illness: evidence from human challenge studies. PLoS One 2016; 11:e0143759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Estes MK, Ettayebi K, Tenge VR, et al. . Human norovirus cultivation in nontransformed stem cell-derived human intestinal enteroid cultures: success and challenges. Viruses 2019; 11:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.