Abstract
To preserve scientific data created by publicly and/or philanthropically funded research projects and to make it ready for exploitation using recent and ongoing advances in advanced and large-scale computational modeling methods, publicly available data must use in common, now-evolving standards for formatting, identifying and annotating should share data. The OpenNeuro.org archive, built first as a repository for magnetic resonance imaging data based on the Brain Imaging Data Structure formatting standards, aims to house and share all types of human neuroimaging data. Here, we present NEMAR.org, a web gateway to OpenNeuro data for human neuroelectromagnetic data. NEMAR allows users to search through, visually explore and assess the quality of shared electroencephalography (EEG), magnetoencephalography and intracranial EEG data and then to directly process selected data using high-performance computing resources of the San Diego Supercomputer Center via the Neuroscience Gateway (nsgportal.org, NSG), a freely available web portal to high-performance computing serving a variety of neuroscientific analysis environments and tools. Combined, OpenNeuro, NEMAR and NSG form an efficient, integrated data, tools and compute resource for human neuroimaging data analysis and meta-analysis.
Database URL: https://nemar.org
Introduction
Although electroencephalography (EEG) was the first functional human brain monitoring modality (1926), EEG data analysis long lagged in adapting new data analysis approaches—both in neurology, where visual pattern recognition applied to the raw scalp signal data is still the dominant approach, and in cognitive neuroscience, where event-related potential averages of individual scalp channel signals, collected from relatively small numbers of participants, long remained the predominant research measure. These methods, however, leave unrevealed much information about brain function contained in the data and also cannot exploit consistencies in complex data that can only be identified in and extracted from large to very large data collections using new statistical and machine learning methods. Sharing neuroelectromagnetic (NEM) data is critical to leveraging public research investment and to supporting rigor and reproducibility in funded research. Several funding bodies require data sharing. Data sharing also allows researchers to use modern research tools to evaluate new data in a new way, by directly comparing it to ever-accumulating stores of shared data collected in related or compatible paradigms.
Because high-density EEG, magnetoencephalography (MEG), and intracranial EEG (iEEG) recordings have no sensors in common, and as important physical differences exist between the dozens of available EEG/iEEG data collection systems and several available MEG systems, a centralized archive of NEM data allowing direct comparison of NEM brain dynamics across studies and modalities has not previously been considered feasible. Furthermore, the broad point-spread function of cortical magnetic flux to scalp coils and the still broader and more variable point-spread function of cortical potentials to scalp electrodes mean that scalp and intracranial NEM recordings have no simply computable relationship. As a result, much valuable convergent information about human brain dynamics contained in the many large and small new and existing NEM data sets, each recorded with care at considerable expense, is at serious risk of being lost to science unless and until the data are integrated into an active integrated data, tools and compute resource (DATCOR) enabling advanced analysis within or across studies.
Here, we report initial results of building NEMAR (nemar.org), a large, publicly available human NEM DATCOR tightly linked to a freely available high-performance computing resource, the Neuroscience Gateway (NSG). We aim to build a widely used and scientifically productive open resource for archiving, sharing and further analyzing and meta-analyzing NEM data.
Data formats
The BIDS format
In the past few years, the magnetic resonance imaging (MRI) community has worked jointly with the International Neuroinformatics Coordinating Facility, to develop community standards for describing and annotating MRI data, the Brain Imaging Data Structure (BIDS) (1). To speed adoption by the brain imaging community, community-driven BIDS standards use common file formats (for MRI, NIFTI, JSON and TSV) and simple directory structures and do not require additional database software. The BIDS MRI standard has now been adopted by several data repositories, including OpenNeuro, FCP INDI, SchizConnect and the Developing Human Connectome Project. The BIDS format for MRI has been responsible in large part for a recent rapid rise in data sharing in the MRI and functional Magnetic Resonance Imaging (fMRI) world (2, 3). BIDS has gained rapid acceptance as an evolving, community-based standard for organizing neuroimaging data to enable efficient data search, advanced processing and data mining. The benefits of the OpenNeuro approach to fMRI data sharing and computation are becoming more apparent, prompting adoption from the NEM data research community. BIDS extensions to MEG data (4) and, more recently, to electrophysiological data including EEG (5) and iEEG data (6) have been developed. Although several small and dedicated NEM data repositories exist (e.g. HeadIT.org, iEEG.org and the OMEGA MEG data archive), these archives do not build around a common data format standard, thwarting cross-modality data co-registration and federation. NEMAR’s support of the emerging BIDS NEM modality standards will allow storage of a wide variety of NEM data collection formats and protocols and make identification, storage and efficient search and retrieval of a wide variety of NEM data possible within a single archive or federated network of archives.
The HED standard
Archived time series data typically require that standardized terms be used to describe the nature of all experimental events of interest identified in the recording session, either during its capture or after the fact, for potential search and re-use in further analysis and meta-analysis. Although BIDS-formatted data sets contain detailed metadata, the BIDS standards in themselves do not constitute a system for adequately describing the timeline of the recording, including occurrences of planned, unplanned or emergent experimental events. The Hierarchical Event Descriptor (HED) system provides a standardized, flexible and readily extensible set of descriptors for experimental events in brain imaging or behavioral experiments (7). HED tagging can be used to describe many types of experiment events in a uniform, but easily extensible and both human- and machine-readable manner. HED has been integrated into BIDS as the de facto standard for describing events in brain imaging experiments. The NEMAR archive is the first open data archive to leverage the use of HED tags to enable data discovery and integration based on experimental events.
Implementation
NEMAR as an extension of the OpenNeuro project
The OpenNeuro archive (8) currently offers more than 644 open neuroimaging datasets from more than 22 000 participants. Its success, to date, reveals that neuroscientists are willing to share their data, although the attractiveness of sharing remains limited as data sharing can require extra work—not currently required by journals or funding agencies—and easy-to-use data meta-analysis and mining tools are not yet available. The OpenNeuro architecture has two main components: web front end and back end database. The custom web front end runs on an Amazon Web Services server. The OpenNeuro data are stored in an Amazon Web Services (AWS) S3 instance, backed by the open-source DataLad data management software. DataLad has a snapshot data mechanism allowing OpenNeuro users to version their data.
The NEMAR project is capitalizing on the ongoing achievements of the OpenNeuro, NSG (9), Open EEGLAB Portal (10) and BIDS standards development projects by creating a community portal to a large and ever-growing archive of human NEM (EEG, MEG and iEEG) brain imaging data, data analysis tools and advanced computational resources. Our overall goal is to support the creation, maintenance, analysis and cross-study mining of human NEM data by seeding and growing a ‘minable’ archive of NEM data deposited in the OpenNeuro resource. After users upload NEM data to OpenNeuro, those data are automatically copied to the San Diego Supercomputer Center (SDSC) storage using the DataLad cloning mechanism. New DataLad snapshots from OpenNeuro are synched daily. NEM-relevant data measures are then automatically computed on the NSG and made available for display to users through the NEMAR web interface.
The NEMAR website (nemar.org), maintained and developed at SDSC, uses the HUBzero web framework, an open-source software platform for building powerful websites that host analytical tools, publish data, share resources, collaborate and build communities in a single web-based ecosystem (13).
NEMAR BIDS-formatting tool
We have released a bids-matlab-tools plug-in (version 6) to export EEG data to BIDS from our popular EEGLAB (11) software environment (sccn.ucsd.edu/eeglab) running on the MATLAB software package (The Mathworks, Inc.). The plug-in allows users to format their EEG raw data using the BIDS-EEG format either via (a) the BIDS export menu item, (b) a pop-up window (Figure 1) or (c) using a scripting interface. As of January 2022, the bids-matlab-tools plug-in, available from the EEGLAB plug-in manager, had been downloaded 844 times.
Figure 1.
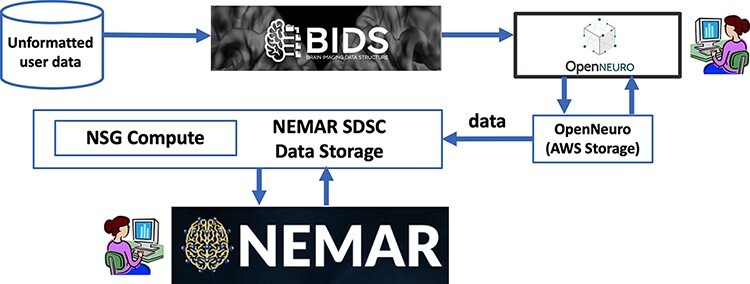
Users first format their data to BIDS and then upload their data to OpenNeuro web interface, which stores it on its AWS back end. The NEMAR SDSC Data Storage back end and then automatically sync/download the data from OpenNeuro. Data statistics and visualizations are precomputed for display by NSG Compute. The NEMAR web interface (bottom box) serves the data to NEMAR users. The NEMAR search engine allows users to search through and select data for analysis for their projects. Data identifiers found on NEMAR (for example, OpenNeuro dataset index ds123456) can then be used in analysis scripts the user sends through the NSG to retrieve and process the selected data without requiring data download and subsequent re-upload.
The plug-in guides users through a series of questions to describe their experiment and populate the BIDS metadata fields, as well as to specify HED tags to record the nature of experiment events and other data timeline structures. Upon completion, the plug-in exports a fully compliant BIDS-formatted dataset that may be uploaded to the OpenNeuro using its web or command line interface, after which the data are automatically made available on NEMAR.
The NEMAR dataset web interface and metadata
For each OpenNeuro NEM dataset, NEMAR automatically extracts and then makes available for display a variety of information extracted from BIDS including the number of EEG channels (and other channel types), the size of the data, the number of files, the BIDS and HED versions, authors, dataset Digital Object Identifier (from OpenNeuro) and other BIDS-related metadata (Figure 2).
Figure 2.
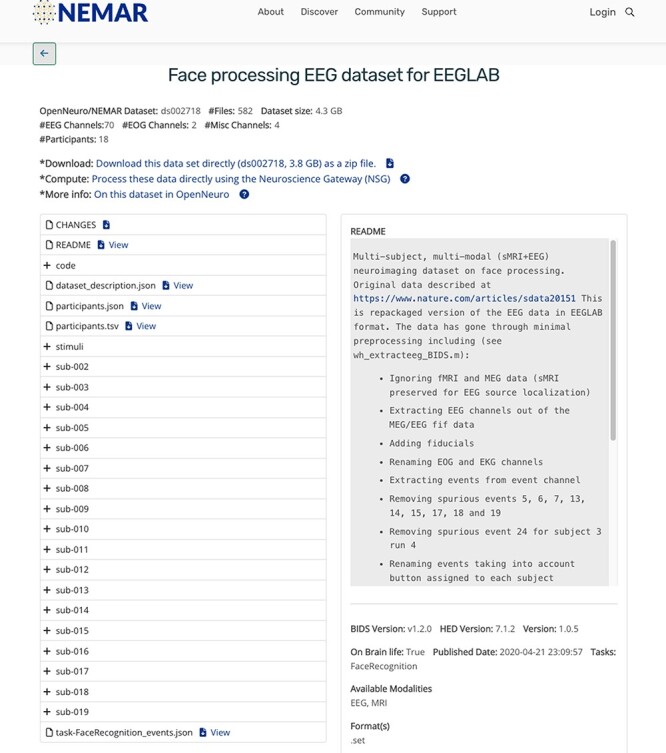
NEMAR dataset interface exposing some of the BIDS-required EEG metadata.
As in OpenNeuro, NEMAR users can explore the BIDS folder content and visualize and/or download data files using the icons adjacent to the data files.
As of February 2022, there were 72 EEG datasets (from 2664 participants) on NEMAR, 22 MEG datasets (from 365 participants) and 12 iEEG datasets (from 202 participants). Of the 72 EEG datasets, 37 use the EEGLAB data format and have likely been formatted using the bids-matlab-tools EEGLAB plug-in (see Figure 3).
Figure 3.
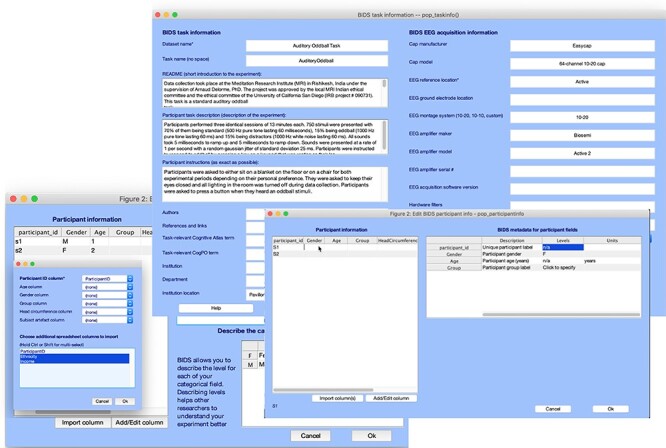
Graphic interface of the bids-matlab-tools plug-in for EEGLAB, a tool to format NEM data in EEGLAB STUDY format to BIDS format for OpenNeuro and NEMAR.
NEMAR data quality pipeline
On NEMAR, we are implementing an automated processing pipeline to compute a variety of data quality metrics, including the percentage of ‘good’ channels and ‘good’ data, and the number of putative brain sources extracted by independent component analysis (ICA), as described in the study by Delorme et al. (12). These measures will be displayed when users click a link next to each dataset (Figure 4).
Figure 4.
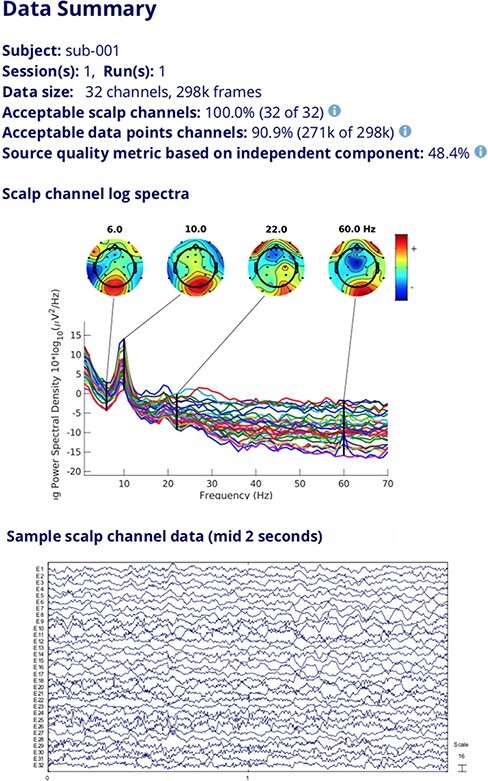
Data quality information, channel log spectra and raw data segment for a selected dataset.
NEMAR data visualization
For each data file of each BIDS dataset, NEMAR also plots the data spectrum and shows a few seconds of the raw data. More data-specific measures will be added in the future. Measures shown by NEMAR are computed running EEGLAB functions via the NSG gateway (see Figure 1). Visualization measures are saved in .SVG vector format and are displayed using native SVG HTML tag capabilities, preserving vector graphic information for adaptive resolution on web browsers.
As part of the data quality pipeline, brain sources are extracted using ICA. We will allow users to visualize ICA components on NEMAR, as well as the putative brain source they correspond to in 3D template head models. Although less accurate than using the subject’s head model, source visualization on template head models may be computed automatically and also allow comparisons across both subjects and BIDS datasets.
NEMAR interface to the NSG
Since 2013, the NSG (9) has been serving the neuroscience community by providing easy access to many software and pipelines running primarily on high-performance computing (HPC) resources provided by the Extreme Science and Engineering Discovery Environment (XSEDE) network that coordinates resources across the NSF-funded supercomputer centers.
NSG has made available high-throughput computing (HTC) and academic cloud computing resources, and neuroscientists of all types have increasingly begun to take advantage of NSG and its easy-to-use interface for running tools and pipelines for processing their data. In addition to XSEDE HPC resources, implemented using singularity, NSG uses backend XSEDE HPC, HTC and cloud resources and makes available a range of computational neuroscience tools including NEURON, PyNN, GENESIS, Brian2, NEST, MOOSE, NetPyNE, BMTK, HNN-Core, etc. NSG has more recently made available environments widely used for human brain imaging research including Freesurfer, MATLAB and Python. In 2017, we ported to NSG our very widely used EEGLAB signal processing environment EEGLAB (11) and its many contributed tools and toolboxes, creating an Open EEGLAB Portal to high-performance data processing resources (10).
The NEMAR gateway project shares the same infrastructure as NSG, and NSG capabilities have been expanded to allow users to run data processing tools and pipelines on NEM data of their own or from the OpenNeuro archive. NEMAR already uses NSG for computing NEM data quality metrics and also allows users to run custom MATLAB and Python scripts on NSG (nsgportal.org) using NEMAR data (Figure 5).
Figure 5.

An example MATLAB script running on NSG using OpenNeuro/NEMAR dataset ds002691. The script, submitted by a user through the NSG interface, is executed on the Expanse supercomputer, for which the NEMAR datasets are immediately accessible via their ‘ds’ index (as below).
Conclusion
The NEMAR (nemar.org) front-end portal to NEM neuroimaging data allows users to search for and optionally explore data submitted to OpenNeuro (openneuro.org) by viewing precomputed data quality metrics and visualized dataset information and then process selected data using the XSEDE high-performance resources in conjunction with the NSG (nsgportal.org) without requiring a data download and subsequent re-upload. NEM data will be stored in both raw and source-resolved formats that will allow source-level data and data measure visualization for user inspection, search, analysis and discovery within and also across studies using a common template brain model—obtaining results largely independent of scalp sensor geometry and modality that can be directly compared to results of other structural and functional neuroimaging research. This feature will also make possible data meta-analyses and large-scale data mining, as tools for this become available. NEMAR thus also serves as an example of an integrated human neuroimaging DATCOR (‘data, tools and compute resource’). As the amount of available shared data grows, integrated computing will become more and more convenient and even necessary. What we here term ‘DATCOR’ seems likely to become the basic unit of open science for advancing research using neuroimaging and other types of publicly shared data.
Acknowledgements
The NEMAR project was supported by NIH grant R24MH120037. The OpenNeuro project is supported by NIH grant R24MH117179. The NSG project is supported by NIH grant U24EB029005, NSF grant 1935749, and NIH grant R01EB023297. We thank Erich Huebner and Claire Stirm of the HubZero team for their support of the NEMAR web interface.
Contributor Information
Arnaud Delorme, Swartz Center of Computational Neuroscience, INC, UCSD, La Jolla, CA 92093-0559, USA; CERCO, CNRS, Paul Sabatier University, Toulouse 31400, France.
Dung Truong, Swartz Center of Computational Neuroscience, INC, UCSD, La Jolla, CA 92093-0559, USA.
Choonhan Youn, San Diego Supercomputer Center, UCSD, La Jolla, CA 92093-0505, USA.
Subhashini Sivagnanam, San Diego Supercomputer Center, UCSD, La Jolla, CA 92093-0505, USA.
Claire Stirm, San Diego Supercomputer Center, UCSD, La Jolla, CA 92093-0505, USA.
Kenneth Yoshimoto, San Diego Supercomputer Center, UCSD, La Jolla, CA 92093-0505, USA.
Russell A Poldrack, Department of Psychology, Stanford University, Stanford, CA 94305, USA.
Amitava Majumdar, San Diego Supercomputer Center, UCSD, La Jolla, CA 92093-0505, USA.
Scott Makeig, Swartz Center of Computational Neuroscience, INC, UCSD, La Jolla, CA 92093-0559, USA.
Data availability
All data provided in the aforementioned database framework are publically available.
Conflict of interest
None declared.
References
- 1. Gorgolewski K.J., Auer T., Calhoun V.D. et al. (2016) The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data, 3, 160044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Poldrack R.A. and Gorgolewski K.J. (2014) Making big data open: data sharing in neuroimaging. Nat. Neurosci., 17, 1510–1517. [DOI] [PubMed] [Google Scholar]
- 3. Poline J.-B., Breeze J.L., Ghosh S. et al. (2012) Data sharing in neuroimaging research. Front. Neuroinform., 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Niso G., Gorgolewski K.J., Bock E. et al. (2018) MEG-BIDS, the brain imaging data structure extended to magnetoencephalography. Sci. Data, 5, 180110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pernet C.R., Appelhoff S., Gorgolewski K.J. et al. (2019) EEG-BIDS, an extension to the brain imaging data structure for electroencephalography. Sci. Data, 6, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holdgraf C., Appelhoff S., Bickel S. et al. (2019) iEEG-BIDS, extending the brain imaging data structure specification to human intracranial electrophysiology. Sci. Data, 6, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Robbins K., Truong D., Appelhoff S. et al. (2021) Capturing the nature of events and event context using hierarchical event descriptors (HED). NeuroImage, 245, 118766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Markiewicz C.J., Gorgolewski K.J., Feingold F. et al. (2021) The OpenNeuro resource for sharing of neuroscience data. eLife, 10, e71774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sivagnanam S., Yoshimoto K., Carnevale N.T. et al. (2018) The neuroscience gateway—enabling large scale modeling and data processing in neuroscience. In: Proceedings of the Practice & Experience in Advanced Research Computing PEARC18. Pittsburgh, PA. pp. 1–7. [Google Scholar]
- 10. Martínez-Cancino R., Delorme A., Truong D. et al. (2021) The open EEGLAB portal interface: high-performance computing with EEGLAB. NeuroImage, 224, 116778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delorme A. and Makeig S. (2004) EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods, 134, 9–21. [DOI] [PubMed] [Google Scholar]
- 12. Delorme A., Truong D., Martinez-Cancino R. et al. (2021) Tools for importing and evaluating BIDS-EEG formatted data. In: 10th International IEEE/EMBS Conference on Neural Engineering (NER). Rome, Italy. pp. 210–213. [Google Scholar]
- 13. McLennan M. and Kennell R. (2010) HUBzero: a platform for dissemination and collaboration in computational science and engineering. In: Computing in Science & Engineering 12.2. pp. 48–53.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data provided in the aforementioned database framework are publically available.


