Figure 1.
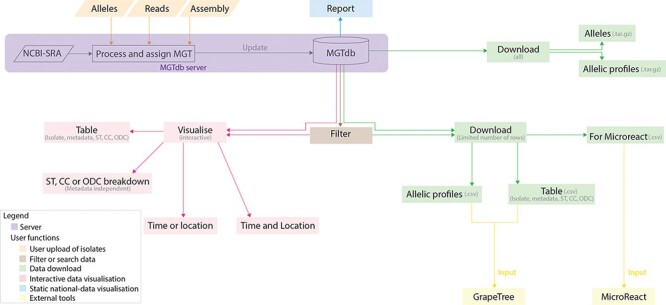
Data visualizations and downloads in MGTdb. The MGTdb database consists of isolates—user-uploaded or those retrieved from NCBI-SRA—assigned the MGT identifiers. In particular, isolates submitted as reads or extracted alleles can be used for typing or querying the database for existing types, whereas isolates submitted as assemblies can only be used for querying the database for existing types. Queried isolates are only visible and downloadable by their uploading user. The MGTdb web service contains features to filter (indicated by brown), visualize (pink and blue) or download (green) these isolates. Indicated by pink: isolates are displayed in a table, either as a complete set or filtered with by metadata, ST, CC and ODC assignments. These data can also be explored via three interactive graphs. The first enables exploring the set of isolates and their MGT assignments in the context of either time or location, the second, in the context of both and the third interactive graph does not use metadata, and enables exploration of isolates and their MGT assignments. Indicated by blue: a report of country-wise or project-wise static summaries of isolates for the last 10 years, at each MGT level, can be generated. Indicated by green: the first 2000 isolates in the displayed table can be exported for use with Microreact—a web-tool which enables phylogeographical and temporal exploration of isolates. The data displayed in the initial table and the allelic profiles for the largest MGT-level STs can also be downloaded. For these two files, the current limits are set to 1 000 000 rows (essentially the entire database) for the tabular data and 10 000 for allelic profiles. The complete set of publicly available isolates, their allelic profiles and allele sequences can be downloaded separately from the organism main page.
