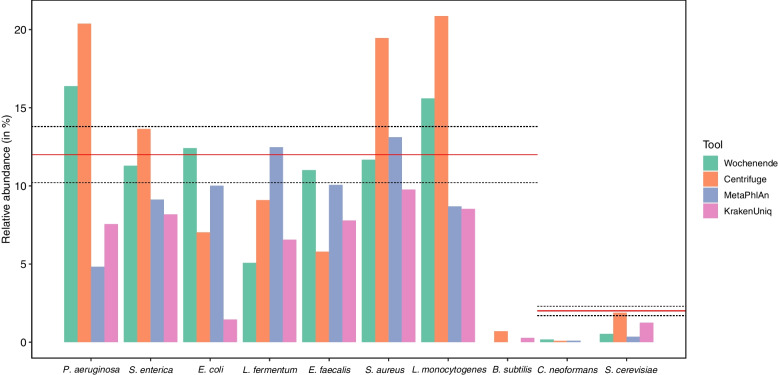
Fig. 2.
Comparison of the performance of four metagenome analysis tools on a single mock community dataset. The mock community of known composition was taken from a published study (SRR11207337) [29]. The red line indicates the expected percentage of each bacterial (12%) and fungal (2%) species in the community. Dashed lines indicate the expected actual deviation from this percentage as indicated by the manufacturer (± 1.8%). All tools perform differently on the various species. Wochenende performs very well, with estimated abundances very close to the expectation for four of the eight bacteria present. MetaPhlAn3 also performs very well, while Centrifuge and KrakenUniq display a degree of over and underprediction, respectively. The bacterium B. subtilis and fungus C. neoformans appear to be present in trace amounts or missing. This is congruent with observations from other mock communities we have seen (Additional File 1, Supplementary Figure S2, other data not shown). Wochenende and MetaPhlAn perform badly at recovering S. cerevisiae reads. In this case Wochenende reference sequences for fungi are in their infancy, and contain too many closely related genomes, which can mask each other