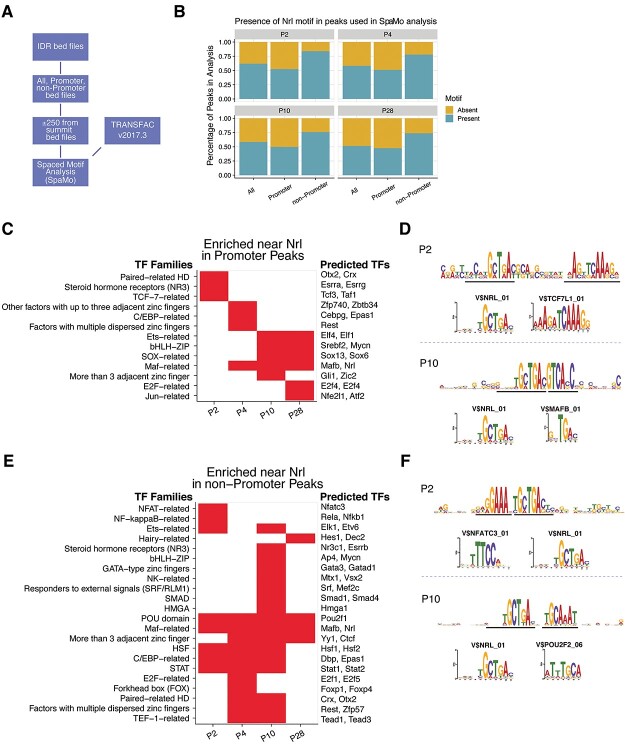
Figure 2.
Temporal transcription factor spaced-motif enrichment. (A) Spaced motif (SpaMo) TF enrichment analysis process overview. NRL motif was used as the primary motif, and all other TRANSFAC vertebrate motifs were considered secondary motifs. (B) Presence of NRL-binding motif in consensus peaks identified in SpaMo analysis. All peaks and peaks annotated as promoter or non-promoter were analyzed at each timepoint. The presence and absence of NRL motif is presented as a percentage of peaks investigated. (C, E) Secondary TF motifs enriched within 150 bases flanking the NRL motif in promoter and non-promoter peaks, respectively. Motif redundancy reduction was achieved by presenting results at the TF family classification level. Individual genes listed on the right of the heatmap are examples of family members expressed in rod photoreceptors. The first gene in the list is the highest-scoring motif in the family, whereas the other gene/s listed are potential candidates based on their ability to bind the same motif and their expression level in the rod photoreceptors. (D, F) Examples of NRL and secondary motif enrichments in promoter and non-promoter peaks, respectively. Enrichment sequence logos of the SpaMo results along with the Transfac motif sequence logos for the highest scoring secondary motifs. Black lines indicate the motif-binding site of enrichment for the primary and secondary motifs. Transfac V$TCF7l1_01 and V$NFATC3_01 motifs are presented from the P2 results (upper panels), whereas V$MAFB_01 and V$POU2F2_06 are shown from the P10 results (lower panels).