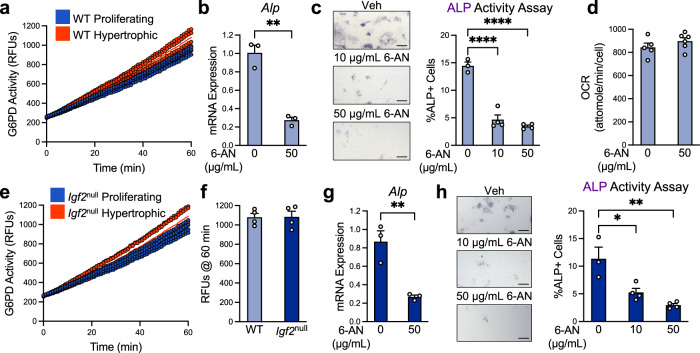
Fig. 6. The pentose phosphate pathway is required for normal hypertrophy in cultured epiphyseal chondrocytes.
a G6PD activity measurements in WT epiphyseal chondrocytes under proliferative and hypertrophic conditions. Representative of n = 3 experiments with 3–4 wells per group. b Gene expression of hypertrophic marker alkaline phosphatase (Alp) as measured by RT-qPCR in WT hypertrophic epiphyseal chondrocytes treated with G6PD inhibitor 6-AN. Representative of n = 2 experiments with 3 wells per group. c WT hypertrophic chondrocytes treated with 6-AN were assessed by staining for alkaline phosphatase (ALP) activity. Representative of n = 3 experiments with 2–4 wells per group. Scale bar: 200 µm. d Baseline OCR measurements of WT epiphyseal chondrocytes under proliferative and hypertrophic conditions and treated with PPP inhibitor 6-AN. Representative of n = 3 experiments with 3–6 wells per group. e G6PD activity in Igf2 null epiphyseal chondrocytes under proliferative and hypertrophic conditions. Representative of n = 3 experiments with 3-4 wells per group. f G6PD activity measurements in WT and Igf2 null epiphyseal chondrocytes under hypertrophic conditions. g Gene expression of hypertrophic marker alkaline phosphatase Alp as measured by RT-qPCR in Igf2 null hypertrophic epiphyseal chondrocytes treated with 6-AN. Representative of n = 2 experiments with 3 wells per group. h Igf2 null hypertrophic chondrocytes treated with 6-AN were assessed by staining for ALP activity. Representative of n = 3 experiments with 2–4 wells per group. Scale bar: 200 µm. Data are presented as individual data points with a line indicating the mean (a, e) or as individual measurements with mean + SEM (b, c, d, f, g, h). Note that the control data (0 µg/mL 6-AN) for b and g are also shown in Fig. 4. Statistical significance was calculated by unpaired two-tailed t-test (b, d, f, g) or one-way ANOVA with Tukey’s test for multiple comparisons (c, h). p-values indicated as *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.