Figure 5. Subset of the ADP-ribosylation reagents can be used to detect and enrich MARylated ssRNA but not NAD+-capped RNA.
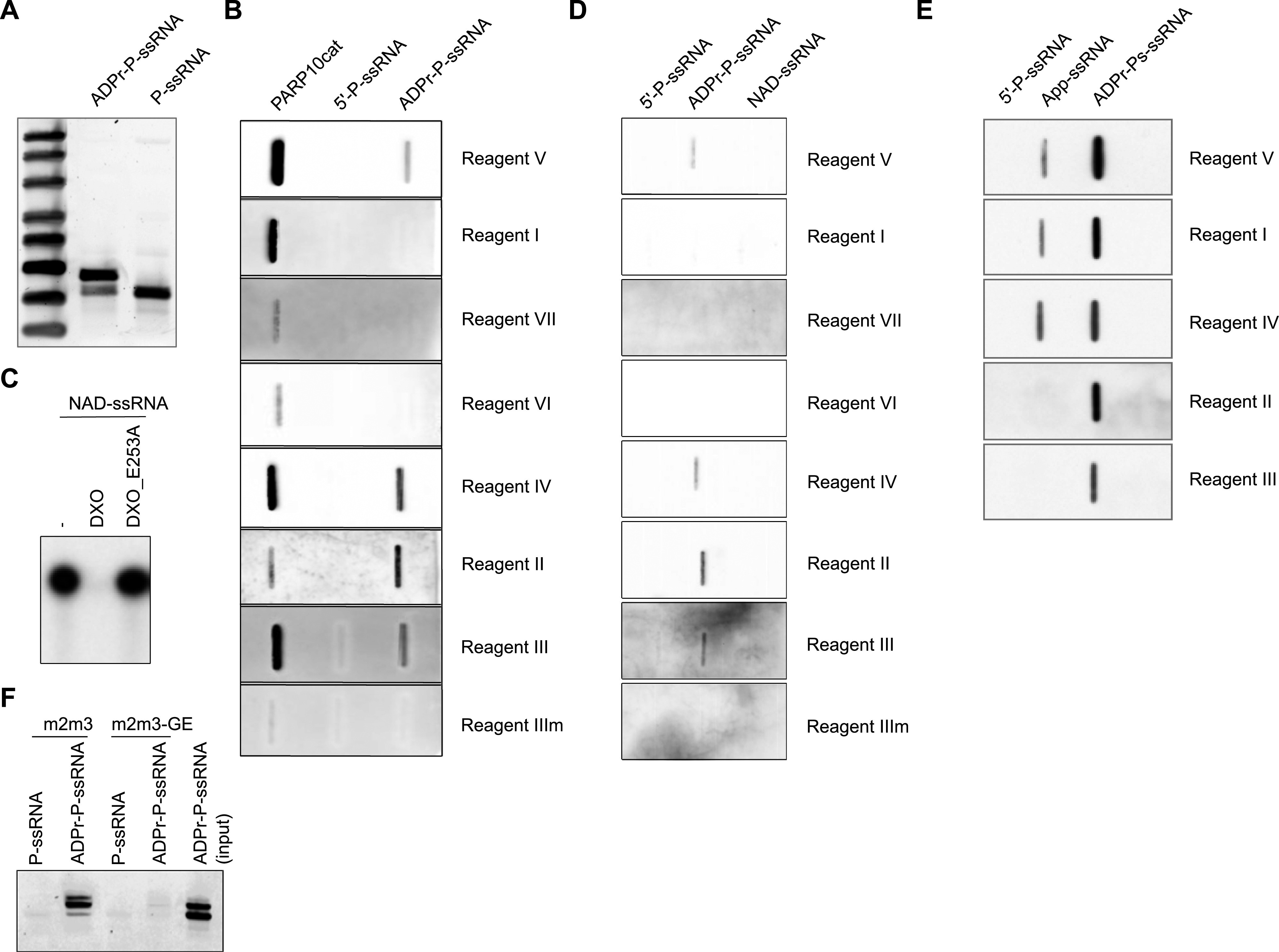
(A) Phosphorylated, single-stranded RNA (P-ssRNA) was ADP-ribosylated with the catalytic domain of PARP10 ranging from amino acids N818–T1025 (PARP10cat), resolved on urea–PAGE, and analysed using SYBR Gold staining. (B) Slot blot was performed with automodified PARP10cat, 5′P-ssRNA, or ADPr-ssRNA. The MARylated ssRNA was treated with proteinase K and purified before slot blotting. The blot was analysed using the indicated detection reagents. (C) Product of an in vitro transcription reaction with 32P-labelled NAD+ was analysed using urea–PAGE and autoradiography. DXO specifically removes the NAD+-cap and degrades the RNA oligo, whereas an inactive mutant is unable to remove the NAD+-cap. (D) As in (B), but now with NAD+-capped RNAs. NAD+-capped RNAs were generated during an in vitro transcription reaction, treated with proteinase K, and purified before analysis by slot blotting. (E) Adenylylated RNAs were slot-blotted and analysed using the indicated reagents. ADP-ribosylated RNA was slot-blotted as a positive control, and phosphorylated RNA, as a negative control. (F) 5′P-ssRNA or ADPr-ssRNA generated as in (A) was incubated with magnetic agarose beads coated with the indicated mPARP14-macro2/3 constructs. After extensive washing, samples were eluted from the beads and analysed on urea–PAGE.
