Figure 6. ADP-ribosylation detection reagents stain different structures depending on the fixation method used.
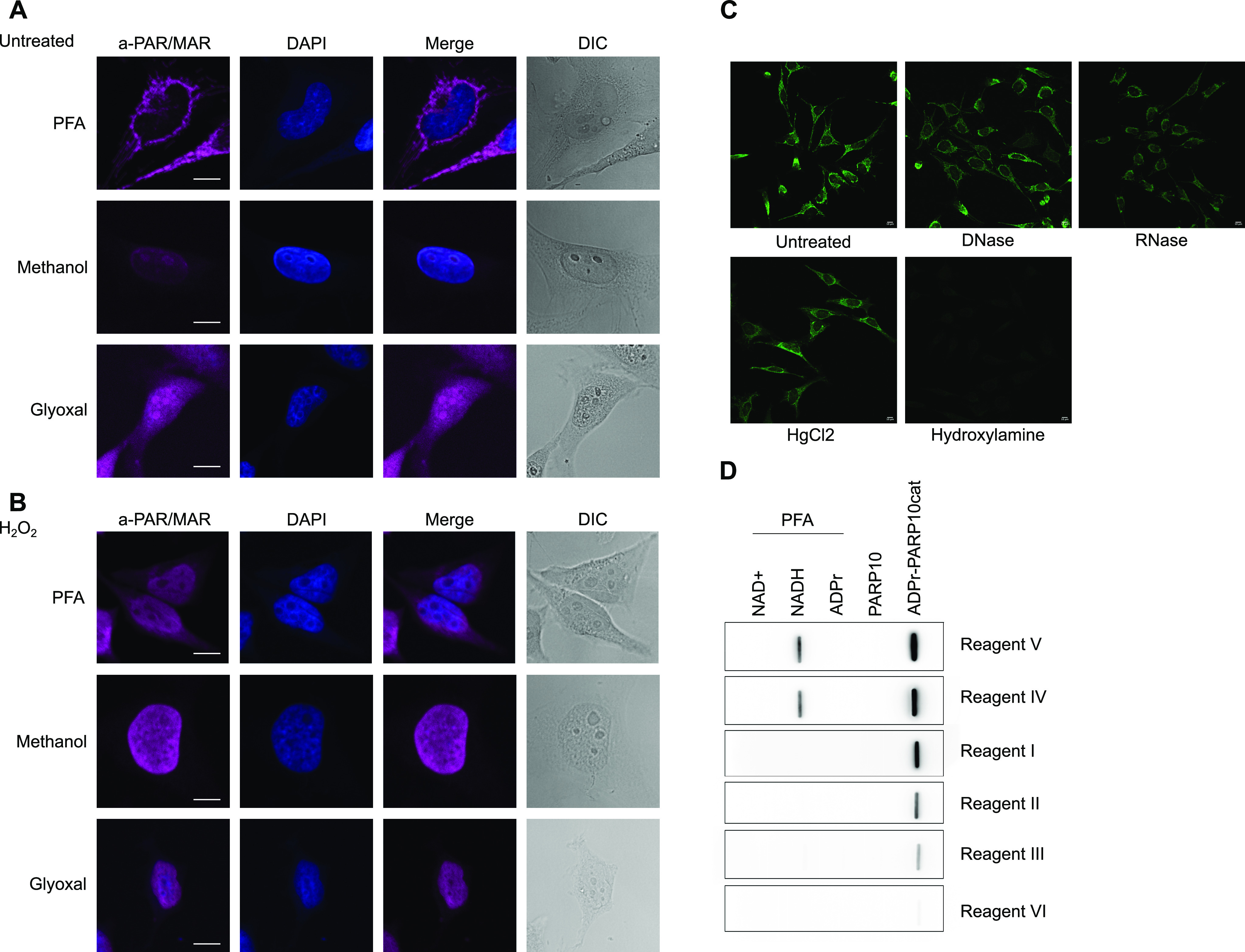
(A) HeLa cells were seeded onto glass coverslips and fixed using PFA, ice-cold methanol, or glyoxal. Reagent V was used to stain the cells, and DAPI was applied to stain the nuclei. (B) As in (A), but before fixation cells were treated with 0.5 mM H2O2 for 10 min. All images were taken using a confocal microscope with identical laser intensity and settings across all samples. (C) HeLa cells were seeded as in (A) and fixed using PFA. Fixed samples were treated with either DNase or RNase, HgCl2, or hydroxylamine. ADP-ribosylation was visualised using Reagent V, and images were taken using confocal microscopy at the same settings for each sample. (D) Indicated metabolites were cross-linked to BSA using PFA and slot-blotted. Blots were incubated with the indicated antibodies and detected using chemiluminescence. Scale bars in (A, B, C) represent 10 µM.
