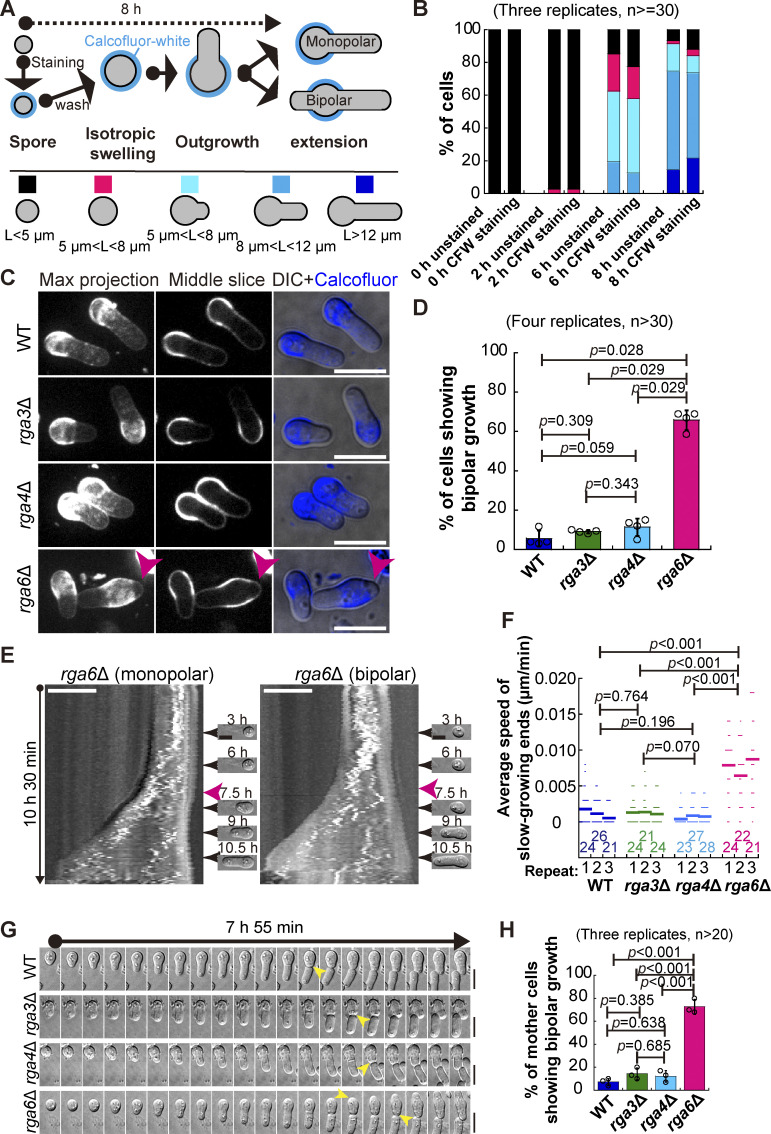
Figure 2.
Characterization of the Cdc42 GTPase-activating proteins Rga3, Rga4, and Rga6 in regulating monopolar growth of germinating cells. (A) Diagrams illustrating the method of using Calcofluor-white staining to determine the growth pattern of germinating cells (i.e., monopolar or bipolar outgrowth). (B) Shown below are the category of spore stages, and quantification in B was based on these categories. L indicates spore length. Average percentage of the indicated categories (in A) of the unstained spores or the spores stained with Calcofluor-white (CFW) at the indicated time after culture in Calcofluor-white–free rich medium. Three independent experiments were carried out, and the number of cells analyzed for each experiment was not less than 30. Additionally, see Fig. S2, C and D, for details. (C) Z-slice (indicated by Middle) and maximum projection images of WT, rga3Δ, rga4Δ, and rga6Δ germinating cells that were stained with Calcofluor-white and cultured in rich medium for 8 h. Pink arrowheads indicate monopolar growth of outgrowing cells. DIC indicates differential interference contrast images. Scale bar, 10 μm. (D) Plot of the percentage of germinating WT, rga3Δ, rga4Δ, and rga6Δ cells (indicated in C) that underwent bipolar growth. Note that four independent experiments were carried out for quantification, and >30 cells were analyzed for each replicate. The top of the column and the error bar are the mean ± SD. The Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. (E) Kymograph graphs of representative rga6Δ spores growing in monopolar and bipolar manners. Images were acquired by time-lapse light microscopy for 10.5 h, and representative DIC images at the indicated timepoint are shown on the right. Pink arrowheads indicate spore outgrowth. Scale bar, 5 μm. (F) Average extension speed of the slow-growing end of WT, rga3Δ, rga4Δ, and rga6Δ spores. The speed was measured using kymograph graphs, as indicated in E, and end displacements taking place during ∼3 h since outgrowth were used for the calculation. All data points from three independent experiments and the number (n) of spores analyzed are shown. Bars represent the mean. For comparisons between groups, the Wilcoxon-Mann-Whitney Rank Sum test was used to calculate the P values. (G) Time-lapse DIC images of WT, rga3Δ, rga4Δ, and rga6Δ spores. Note that the pattern of monopolar growth of WT, rga3Δ, and rga4Δ parental cells, but not rga6Δ parental cells, was maintained after the first cell. Yellow arrowheads indicate outgrowth. Scale bar, 10 μm. (H) Quantification of the percentage of WT, rga3Δ, rga4Δ, and rga6Δ parental cells showing bipolar outgrowth after the first cell division. Three independent experiments were carried out, and the number of cells analyzed for each experiment was not less than 20. The top of the column and the error bar are the mean ± SD. One-way ANOVA with Tukey HSD post hoc test was used to calculate the P values.