Figure 1. Temperature-sensitive ole1 mutants are compromised in cell growth and accumulate saturated lipids.
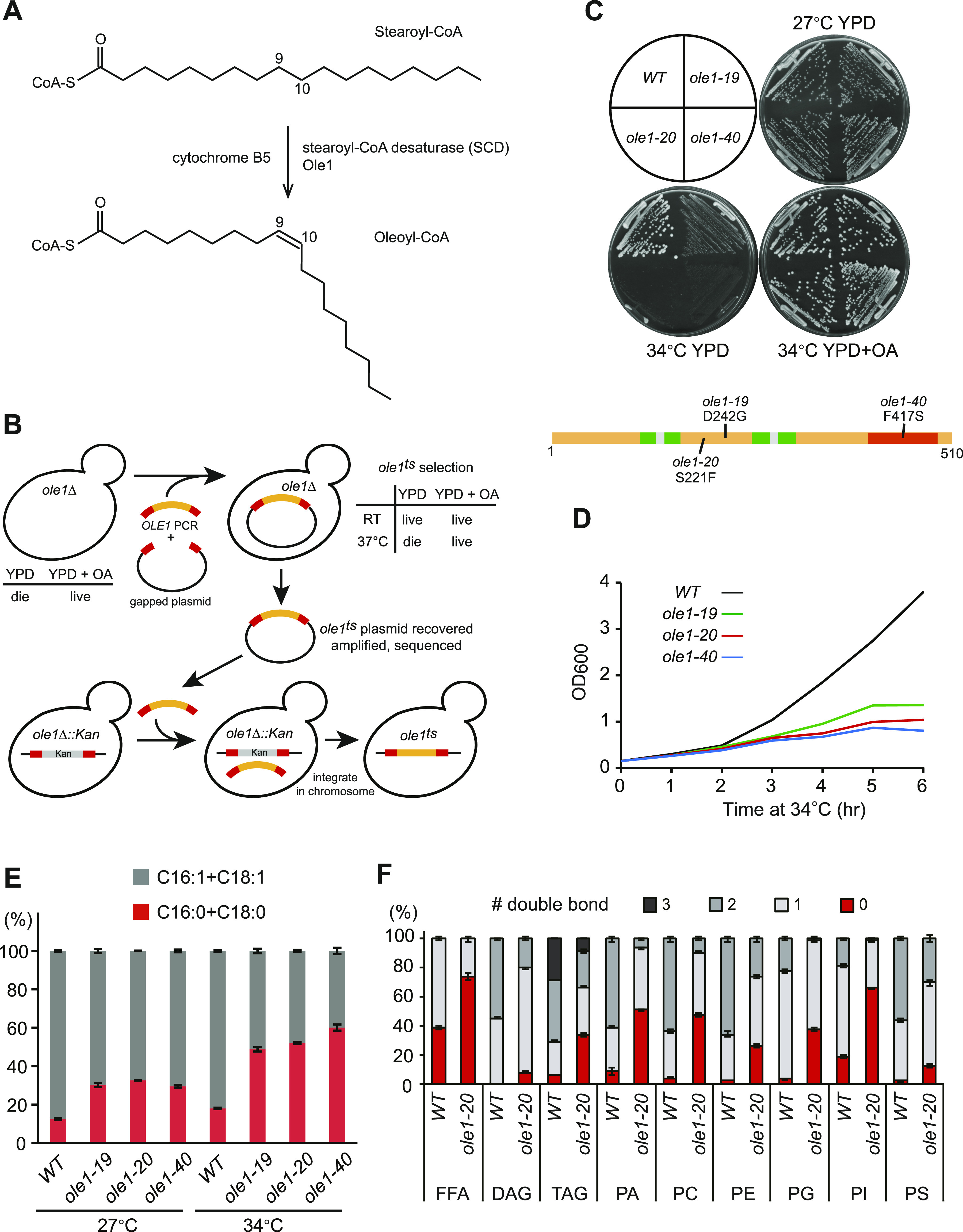
(A) Conversion of stearoyl-CoA to oleoyl-CoA by stearoyl-CoA desaturase, Ole1 in yeast, and cofactor cytochrome B5. (B) Schematic diagram of the screen for temperature-sensitive ole1 mutants. RT, room temperature (∼25–27°C). OA, oleic acid. (C) Growth of three temperature-sensitive ole1 mutants on indicated plates at 27°C or 34°C. The diagram shows the domain structure of Ole1 and the mutation sites of three ole1 alleles. Green, transmembrane domains; red, cytochrome B5 domain; orange, cytoplasmic region; and gray, ER luminal region. (D) Growth rate of WT and ole1 mutants at 34°C. (E) GC-MS analysis of total fatty acids from cells grown at 27°C or shifted to 34°C for 5 h. The percentage of the most abundant SFAs (C16:0 and C18:0) and UFAs (C16:1 and C18:1) is presented as the mean ± SD from three biological repeats. (F) LC-MS analysis of free fatty acids and glycerolipids from cells grown at 34°C for 5 h. The percentage of lipids containing indicated numbers of double bonds for each lipid class is shown as the mean ± SD from three biological repeats. DAG, diacylglycerol; TAG, triacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; and PS, phosphatidylserine.
