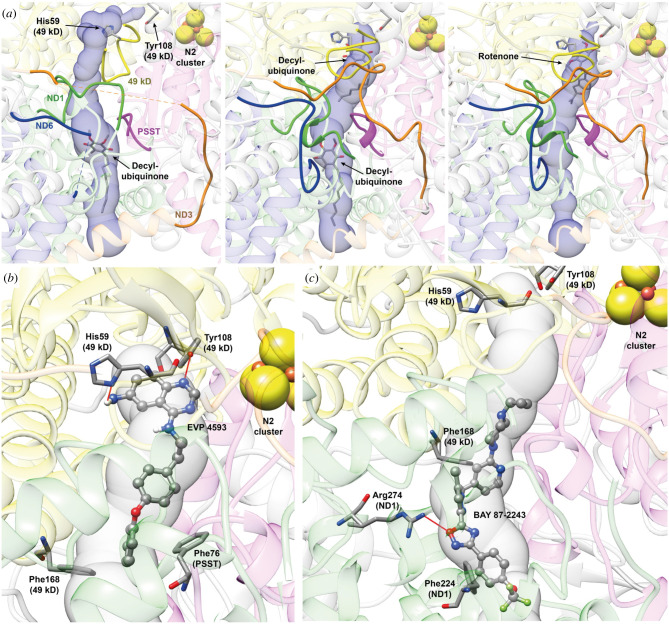
Figure 5.
Molecular docking reveals different binding sites for EVP 4593 and BAY 87-2243 in ovine respiratory CI. (a) Q-site channel departing from Tyr108 (49 kDa) calculated using the CI structure in the initial step of the catalytic cycle (PDB ID 6ZKI, left panel), in the close conformation corresponding to the quinone reduction step (PDB ID 6ZKC, central panel) and in the rotenone inhibited structure (PDB ID 6ZKK, right panel). Protein backbones are reported in transparent cartoons coloured in grey, except for the 49 kDa, PSST, ND1, ND3 and ND6 subunits that are in yellow, purple, green, orange and blue, respectively. The loops cited by Kampjut et al. [44] are in solid ribbons. Decyl-ubiquinone and rotenone found in the structures are reported in sticks coloured according to the atom type, as well as His59 (49 kDa) and Tyr108 (49 kDa). The N2 [4Fe4S] cluster is shown as spheres coloured according to the atom type. The Q-site channel is reported in transparent light blue, to show the position of the ligand in the cavity. (b,c) Detail of the best docking poses for EVP 4593 (b) and BAY 87-2243 (c) calculated on the CI structure in the close conformation corresponding to the quinone reduction step (PDB ID 6ZKC). Inhibitors are shown in ball-and-stick coloured according to the atom type, while CI residues forming specific interaction with the ligands are in stick coloured accordingly to atom type. H-bonds are shown using red lines.