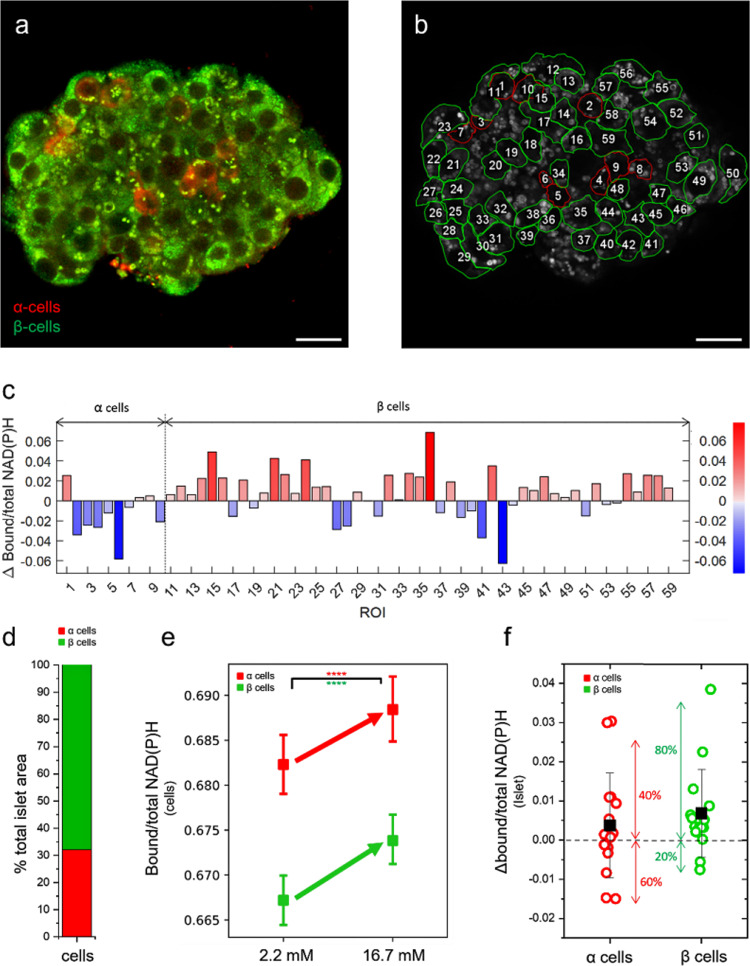
Fig. 3. From FLIM imaging to single-cell metabolism.
a Immediately after 16.7 mM glucose FLIM imaging, cells are fixed to preserve their shape and structure and then incubated with anti-glucagon and anti-insulin antibodies which in turn recognize α (red) and β -cells (green). b Comparison of 2.2 and 16.7 mM FLIM images with immunofluorescence image allow us to recognize cell borders and identity. At this point, single-cell segmentation occurs by tracing ROIs on the two FLIM images (here reported only the 2.2 mM as representative image). Each segmented cell has a unique and equal ROI number in both 2.2 and 16.7 mM FLIM images. Then, single-cell barycenter are calculated, converted in Bound/total NAD(P)H ratio and their difference between 2.2 and 16.7 mM state determines the single-cell metabolic shifts (c). Each cell ROI represents a single cell with red bars indicating bound-shift and blue ones free-shift. d Considering all the segmented cells, the relative abundance of α (~30%) and β cells (~70%) is in keeping with literature3 and their average response is bound-shifted for both cell types (e), in apparent contrast with literature. f If single-cell metabolic shifts are averaged at islet level, α cells (n = 312 cells from 15 islets obtained by 4 different donors) show shift towards free NAD(P)H in 60% of islets with the remaining 40% shifting towards bound NAD(P)H; by contrast, for what concerns β cells (n = 654 cells from 15 islets obtained by 4 different donors), approximately 80% of the islets display a metabolic shift towards bound NAD(P)H and only the remaining 20% towards free NAD(P)H. Statistical difference obtained using Paired Samples Wilcoxon Test, with significance α = 0.05 (****P < 0.0001). Scale bar: 20 μm.