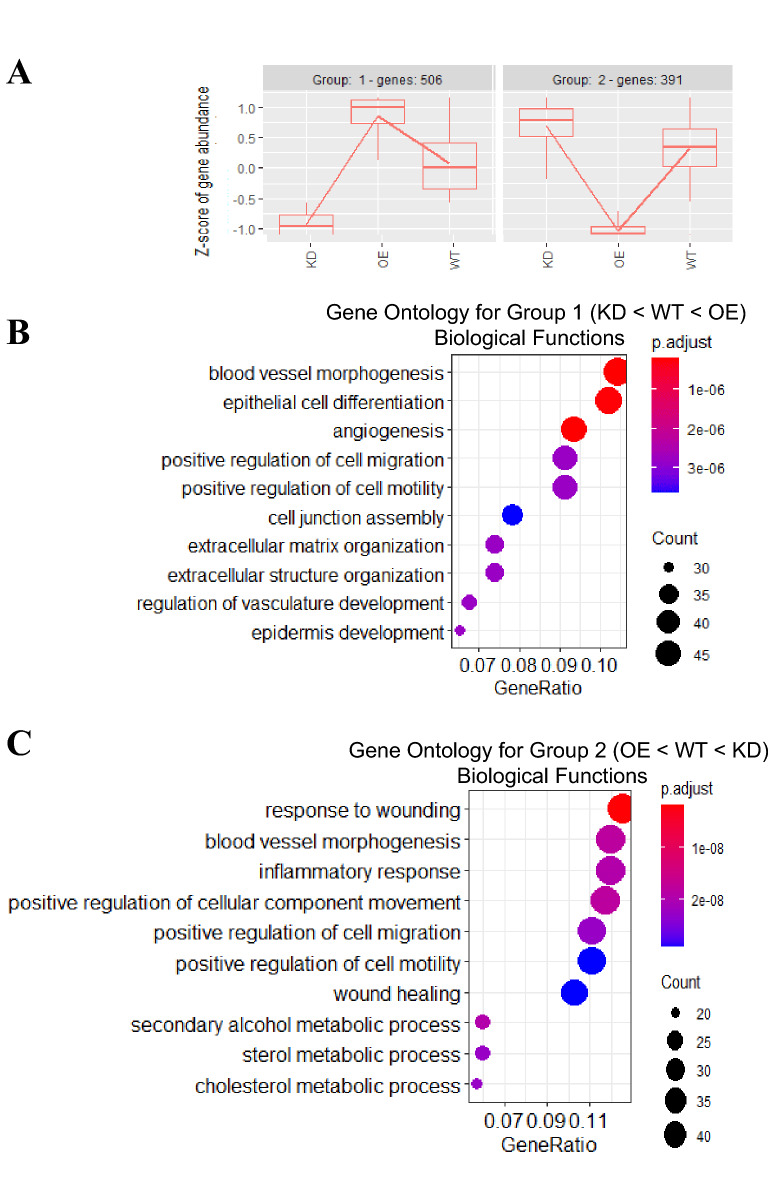
Figure 3.
Transcriptome patterns and properties of genes impacted by EMP2 modification. Changes in gene expression across wild type (WT), EMP2 overexpressing (OE), and EMP2 knock down (KD) ARPE-19 cells were queried by the likelihood ratio test. (A) Hierarchical clustering analysis of the top 1000 significant genes (FDR < 0.01) by the degPatterns tool within the DEGreport R package (version 1.26.0) reveals four distinct expression profile groups. (B, C) Enriched biological property gene ontology (GO) terms for group 1 and group 2 cluster genes, respectively. The top 10 GO terms ordered by gene ratio (number of GO term-related genes divided by total number of significant genes) identify angiogenesis and vasculature-related terms as significantly enriched for group 1 genes. Dot color represents the Benjamini–Hochberg adjusted p value for each enriched GO term. All samples were performed in triplicate.