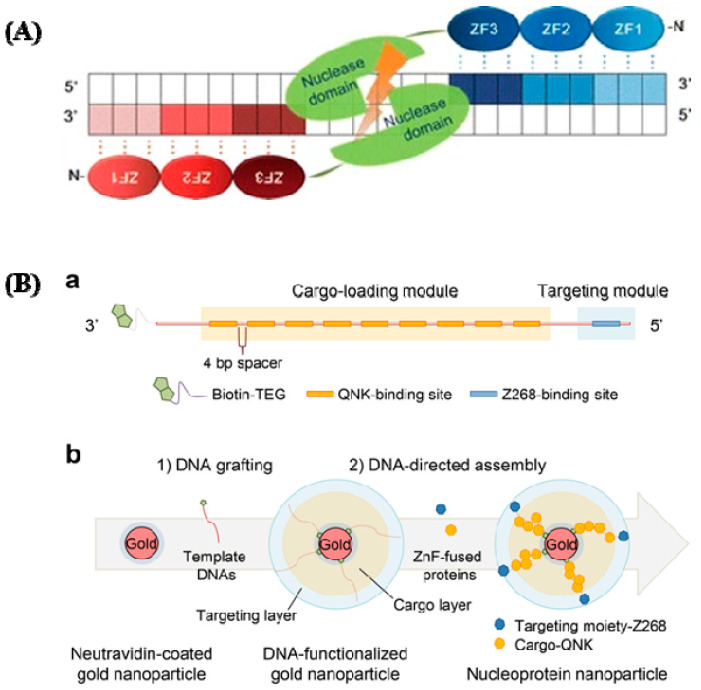
Figure 3.
(A) ZFNs have a specific structure. ZFNs linked to their target locations are depicted schematically. This is a well-designed ZF DNA domain fused to the FokI type IIS restriction enzyme’s DNA cleavage domain that forms nucleases. Each ZFN protein has three ZFs (shaded ovals) that recognize a target sequence in the DNA (shaded boxes). The binding of two ZFNs on target DNA with a spacer (5/6 bp) in the center causes the ZFN nuclease domains to dimerize and the target DNA to be cleaved (reprinted with permission from [51]). (B) DNA and zinc fingers are used to program the construction of nucleoprotein nanoparticles. (a) Template modular DNA design. A biotin molecule was added at the 3′ end of the template DNA with a TEG linker for grafting template DNA onto AuNPs. To eliminate steric hindrance, multiple binding sites for QNK-QNK-RHR (QNK) were spatially organized with a four-base-pair spacer. (b) Sequential construction of nucleoprotein nanoparticles is depicted schematically (NNPs) (reprinted with permission from [52]).