Figure 6.
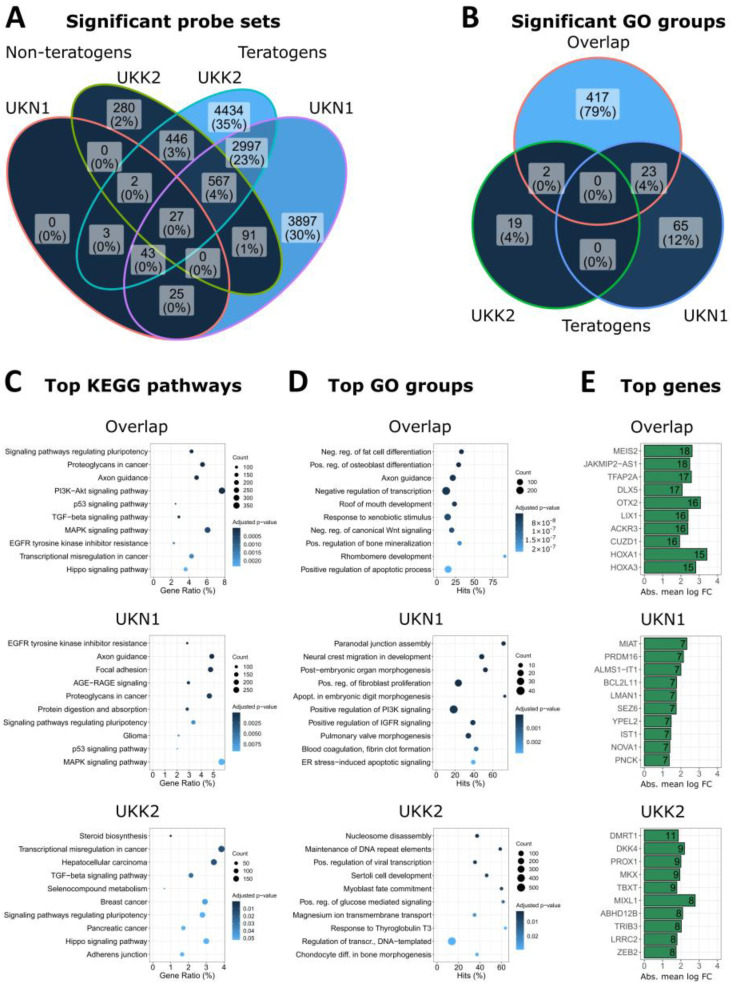
Biological interpretation and comparison of genes differentially expressed in the UKN1 and UKK2 test after exposure of hiPSC to teratogens at 20-fold Cmax. (A) Number of significant probe sets (log2 fold change >1; adjusted p-value < 0.05) induced by non-teratogens and teratogens at the 20-fold Cmax (including also 10-fold Cmax carbamazepine and 1.67-fold Cmax VPA). The following gene sets were defined: ‘Overlap:’ SPS that were deregulated by teratogens in UKN1 and UKK2 (3634 SPS); ‘UKN1′ and ‘UKK2:’ SPS that were deregulated by teratogens exclusively in UKN1 (4013 SPS) and UKK2 (4885 SPS), respectively. (B) Number of significantly (adj. p-value < 0.05) overrepresented GO groups in the gene sets ‘Overlap,’ ‘UKN1,’ and ‘UKK2.’ (C) KEGG pathway enrichment analysis of the gene sets ‘Overlap,’ ‘UKN1,’ and ‘UKK2.’ The ten KEGG pathways with the lowest adj. p-values are given. Full names and complete KEGG-pathway lists are given in the Supplementary Excel-file 4. ‘‘Count:’’ number of significant genes from A linked to the KEGG pathway. ‘‘Gene Ratio:’’ percentage of significant genes associated with the pathway compared to the number of all significant genes associated with any pathway. (D) The ten GO groups with the lowest adj. p-values from all significantly (adj. p-value <0.05) overrepresented GO groups in each gene set. The following adjustments applied here: ‘Overlap’ included all GO groups encompassed by the overlap-circle in B (442 GO groups); ‘UKN1′ (65 GO groups) and ‘UKK2′ (19 GO groups) only considered remained GO groups. The names of the GO groups were shortened. Full names and complete GO group lists can be found in the Supplementary Excel-file 5. ‘‘Count:’’ Number of significant genes from A linked to the GO group. ‘‘Hits:’’ percentage of significant genes compared to all genes assigned to the GO group. (E) Top-10 genes deregulated by teratogens within each gene set. The number in the bar indicates the number of compounds that deregulated the specific gene. The absolute mean log fold-change of each gene is given on the x-axis. A comprehensive gene list is provided in the Supplementary Excel-file 6.