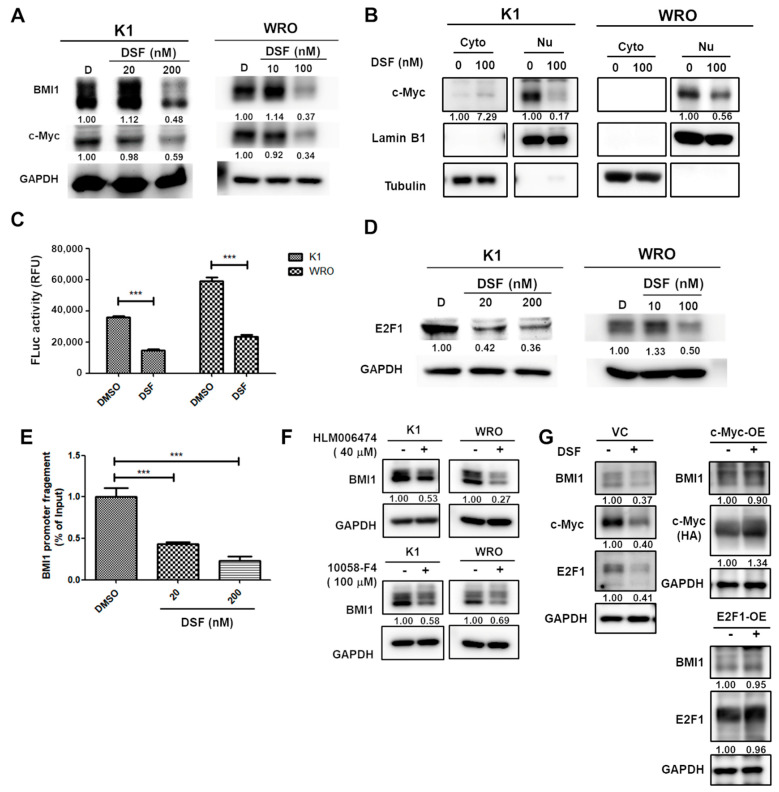
Figure 5.
DSF/copper reduces c-Myc and E2F1 activation in DTC cells. (A,B) Total cellular proteins (A) or the cytoplasmic (Cyto)/nuclear (Nu) fractions (B) of K1 or WRO cells after DSF/copper treatment were extracted and the c-Myc protein was determined by Western blot. Lamin B1 or tubulin in (B) was used as the marker for nuclear or cytoplasmic proteins, respectively. (C) The E2F1 protein expression in total cellular proteins from DSF/copper treated K1 or WRO cells was determined by Western blot. (D) pMyc-Luc and pRL vectors were mixed as a ratio of 100:1 and were transfected into K1 or WRO cells for 24 h followed by treatment of 100 nM DSF in presence of 1 μM CuCl2 for further 48 h. After harvesting the total cell lysates with passive lysis buffer, firefly luciferase or activities were then measured, and data were normalized with Renilla luciferase activity of each sample. ***, p < 0.001. (E) Chromatin DNA of DSF/copper treated K1 cells was extracted and performed immunoprecipitation with anti-E2F1 antibody. The BMI1 promoter DNA fragments within precipitated chromatins were then quantitated by real-time PCR method. The data were presented as relative percentage of input DNA. ***, p < 0.001. (F) K1 or WRO cells were treated with HLM006474 or 10058-F4 at 40 μM or 100 μM, respectively, for 48 h and the protein expression of BMI1 was determined by Western blot. The inserted numbers indicated relative expression levels compared to non-treated samples. (G) WRO cells were transfected with pCMV3 (VC) or vectors carrying cDNA of c-Myc (c-Myc-OE) or E2F1 (E2F1-OE) for 24 h followed by treatment with 100 nM DSF in presence of 1 μM CuCl2 for further 48 h. The protein expressions of BMI1, c-Myc, or E2F1 were determined by Western blot. For c-Myc overexpression, exogenous c-Myc was tagged with hemagglutinin (HA) and was detected by anti-HA antibody. The inserted numbers indicated relative expression levels compared to non-DSF treatment samples.