Figure 1.
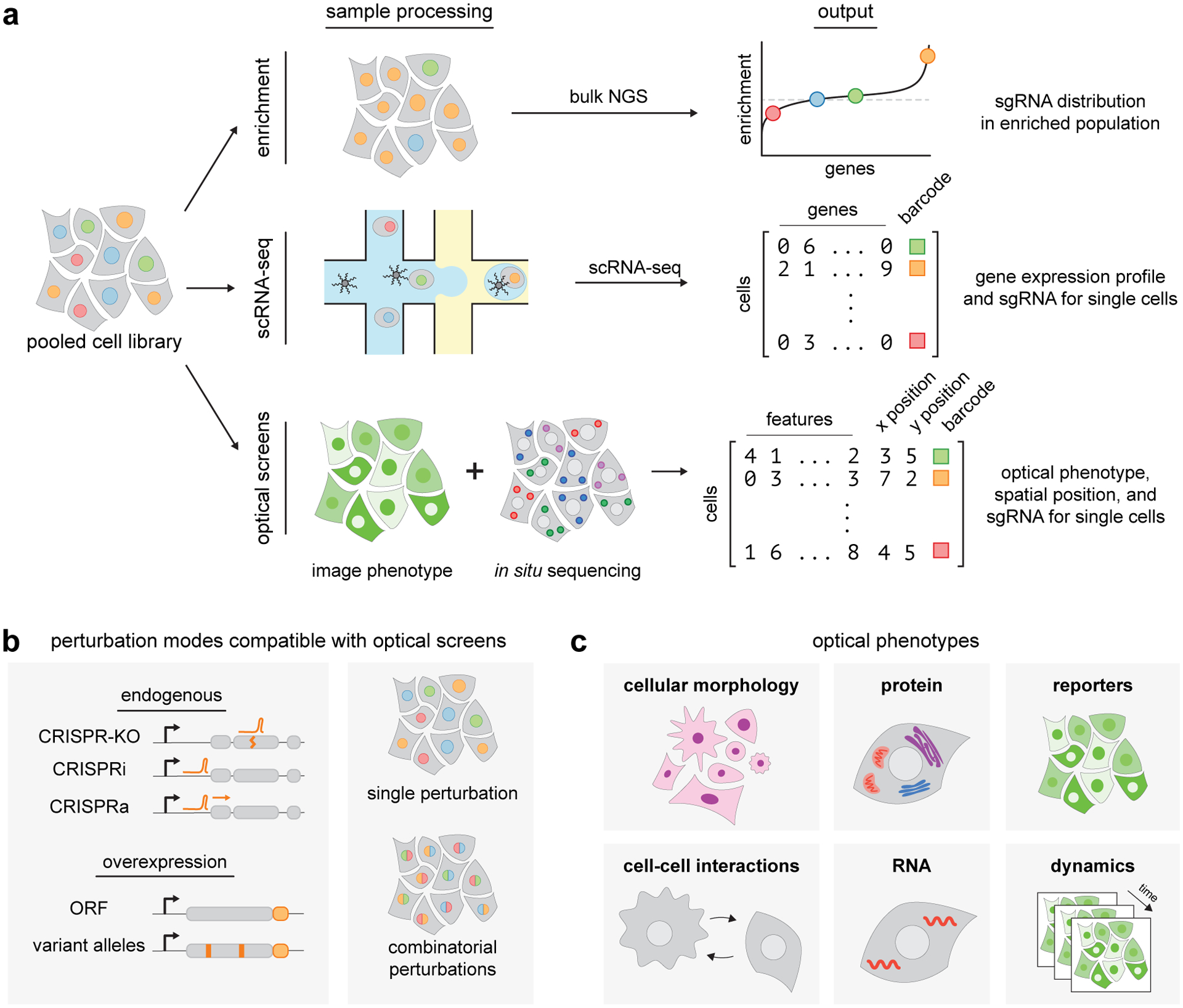
Pooled screening approaches and applications of optical pooled screens. (a) In pooled screening, a population of cells is subjected to a library of genetic perturbations, such as guide RNAs for CRISPR screens. Enrichment, single-cell profiling, and optical-based assays are three common approaches for phenotypic readout. Enrichment-based screens determine population-level changes in perturbation abundance by bulk next-generation sequencing (NGS) following an applied selection. Single-cell profiling and optical screens do not require an enrichment step and instead rely on information-rich phenotypic measurements. Single-cell assays pair perturbation barcodes to a cell phenotype at single-cell resolution, such as cell transcriptome for single-cell RNA sequencing-based screens. Through in situ sequencing, optical pooled screens pair image-based phenotypes with perturbation barcodes, also at single-cell resolution. (b) Optical screens are compatible with multiple perturbation modalities, including CRISPR-based perturbations of endogenous genomic loci and exogenous overexpression of barcoded transgenes. The single-cell readout enables both single and combinatorial perturbation screens. (c) Optical screens enable rich phenotypic measurements, including cellular morphology, cell-cell interactions, dynamic behaviors, and abundance and localization of endogenous protein and RNA molecules and exogenous reporters.
