Figure 2.
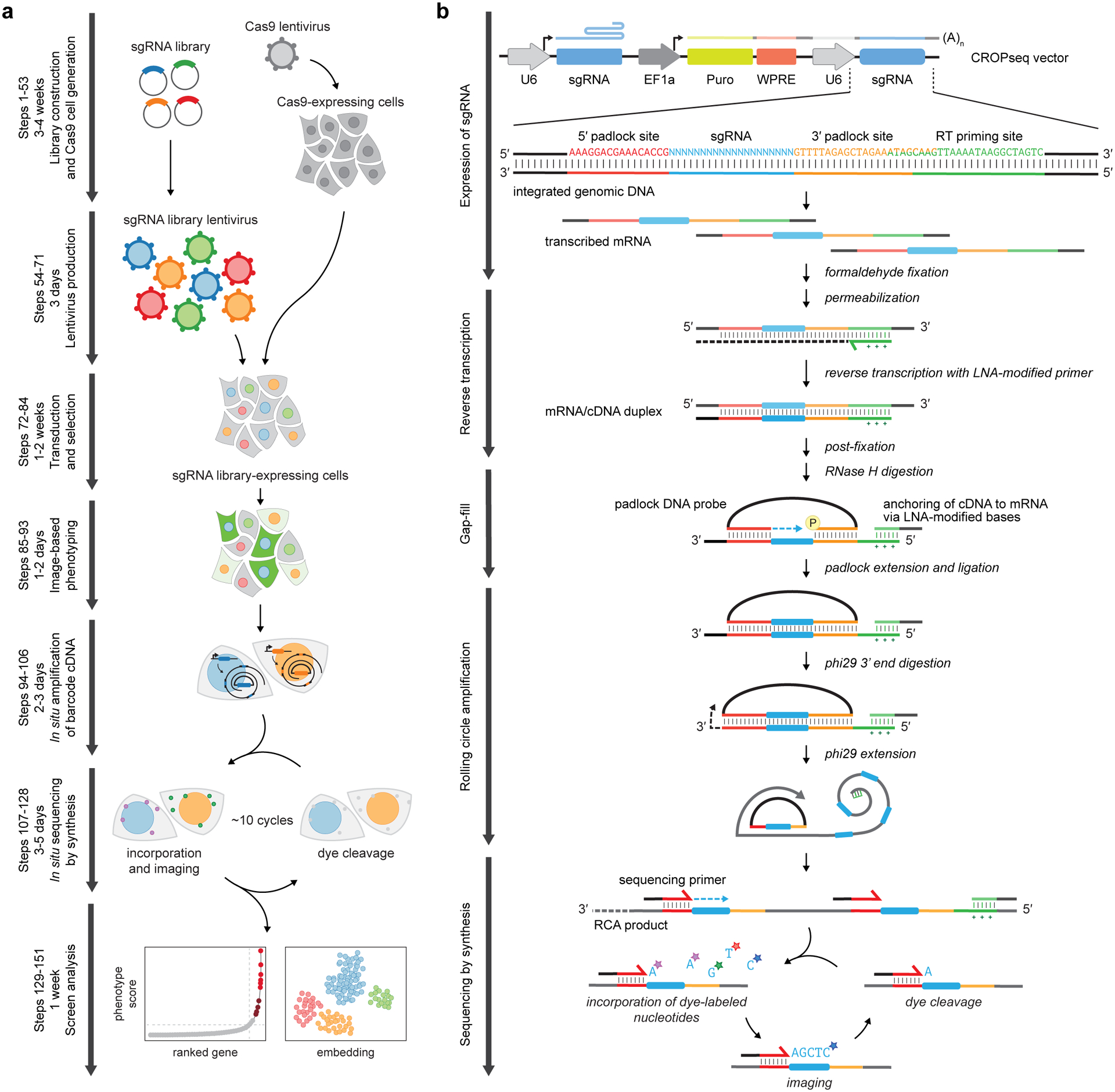
Overview of optical pooled screening. (a) Experimental workflow. First, a pooled sgRNA library is designed, packaged into lentivirus, and delivered into Cas9-expressing cells. A live-cell or fixed-cell imaging assay is performed to generate an optical phenotypic profile of individual cells. The sgRNA spacer sequences in each cell are then amplified and read out by in situ sequencing by synthesis (SBS), consisting of cycles of dye incorporation, imaging, and cleavage. Finally, sgRNA-encoded perturbations are mapped to phenotypic scores at the single-cell level, with candidate genes identified through various statistical approaches. (b) Schematic of the in situ SBS process. The sgRNA is expressed as a polyadenylated mRNA transcript from an integrated copy of the CROPseq vector. After fixation and permeabilization, a locked nucleic acid (LNA)-modified primer is used to reverse transcribe a cDNA copy of the sgRNA sequence. After glutaraldehyde and formaldehyde post-fixation, the mRNA is digested and a padlock probe is hybridized to cDNA regions flanking the sgRNA sequence. The padlock probe is then extended and ligated to copy the sgRNA sequence into a single-stranded circularized DNA. This circularized DNA serves as a template for rolling circle amplification with Phi29 polymerase, the amplified product of which contains tandem repeats of the sgRNA spacer sequence. These sequences are read out by successive cycles of SBS.
