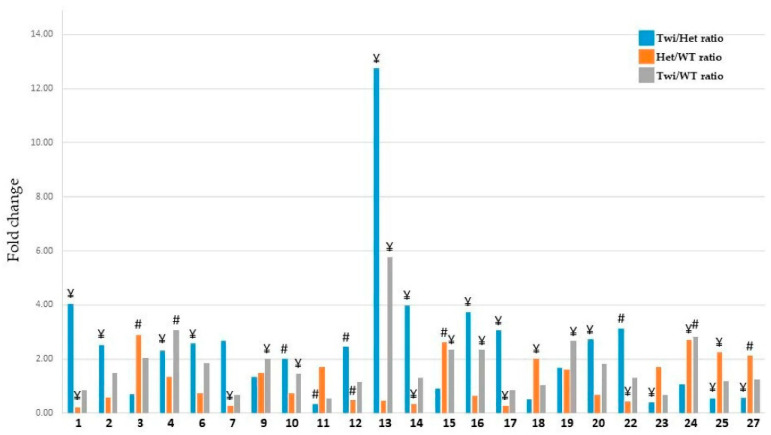
Figure 2.
Histogram representing the %Vol ratios of Twi vs. Het (blue bar), Het vs. WT (orange bar), and Twi vs. WT (grey bar) as reported in Table 1. The numbers in the x axis correspond to those of differentially abundant protein spots identified by mass spectrometry, for more clarity: 1: Ubiquitin carboxyl-terminal hydrolase 5; 2: Transitional endoplasmic reticulum ATPase; 3: Aconitate hydratase, mitochondrial; 4: Aconitate hydratase, mitochondrial; 6: Aconitate hydratase, mitochondrial; 7: Albumin; 9: Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial; 10: Dihydropyrimidinase-related protein 2; 11: Lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial; 12: 2′,3′-cyclic-nucleotide 3′-phosphodiesterase; 13: Vimentin; 14: ATP synthase subunit beta, mitochondrial; 15: Protein NDRG1; 16: NSFL1 cofactor p47; 17: Tropomodulin-2; 18: 60S acidic ribosomal protein P0; 19: Beta-soluble NSF attachment protein; 20: Creatine kinase B-type; 22: Receptor of activated protein C kinase 1; 23: Protein-L-isoaspartate(D-aspartate) O-methyltransferase; 24: Translationally-controlled tumor protein; 25: Transgelin-3; 27: Myelin basic protein. # and ¥ indicate statistical relevance, i.e. p ≤ 0.01 and 0.01 < p ≤ 0.05, respectively.