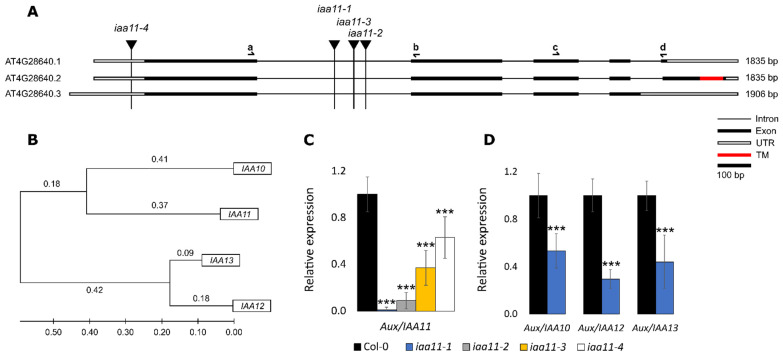
Figure 1.
Identification of T-DNA mutants in Aux/IAA11 gene, and phylogenetic analysis of Aux/IAAs. (A) Splicing variants of Aux/IAA11 gene. Black vertical lines represent the insertion of T-DNA in iaa11-1, iaa11-2, iaa11-3, and iaa11-4 mutant lines. The red rectangle indicates the localization of the transmembrane (TM) domain in Aux/IAA11.2. Black half arrows illustrate binding sites of the primers used for qPCR. (B) Cladogram of protein similarities. Aux/IAA11 clade was prepared using the ClustalW alignment algorithm and the maximum likelihood statistical method in Molecular Evolutionary Genetics Analysis (MEGA) 11 software [60]. (C) Relative expression of Aux/IAA11 in tested mutant lines. (D) Relative expression of Aux/IAA10, Aux/IAA12, and Aux/IAA13 in iaa11-1 mutant background. Relative gene expressions were calculated and normalized with UBIQUITIN-PROTEIN LIGASE 7 (UPL7) and YELLOW-LEAF-SPECIFIC GENE 8 (YLS8) as references, using the Relative Expression Software Tool (REST 2009) (n = 6, *** p < 0.001). Error bars represent the standard deviation.