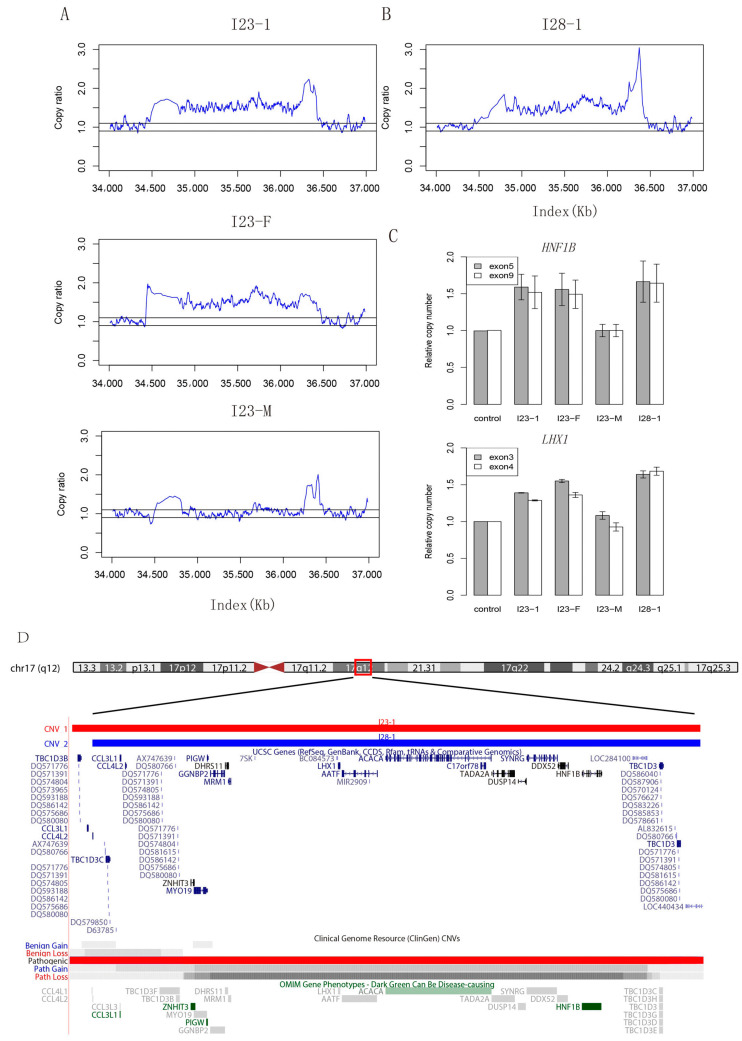
Figure 1.
Copy number variants and validation of patients. (A) Copy ratio of mapped reads on 17q12 in family 23. Read depth shows a heterozygous fragment duplication in patient I23-1 and her father, while her mother has a normal structure in this region. I23-1: patient (described as I23 in text); I23-F: patient’s father; I23-M: patient’s mother. (B) Copy ratio of mapped reads on 17q12 in patient I28. Read depth shows a fragment duplication in patient I28-1 (patient I28 in the text). (C) Bar plot of q-PCR validation results for HNF1B and LHX1. Primers were designed for two exons of these two genes. The height of a bar indicates the relative copy ratio compared with the control cell line (GM20764) having a normal diploid genome in these regions. (D) Integrative genomics view of copy number variants on 17q12 and affected genes. Genome browser of duplicated fragments detected in I23-1 and I28-1, along with affected genes within this region. Records of gain and loss of this fragment in public database are also shown at the bottom.