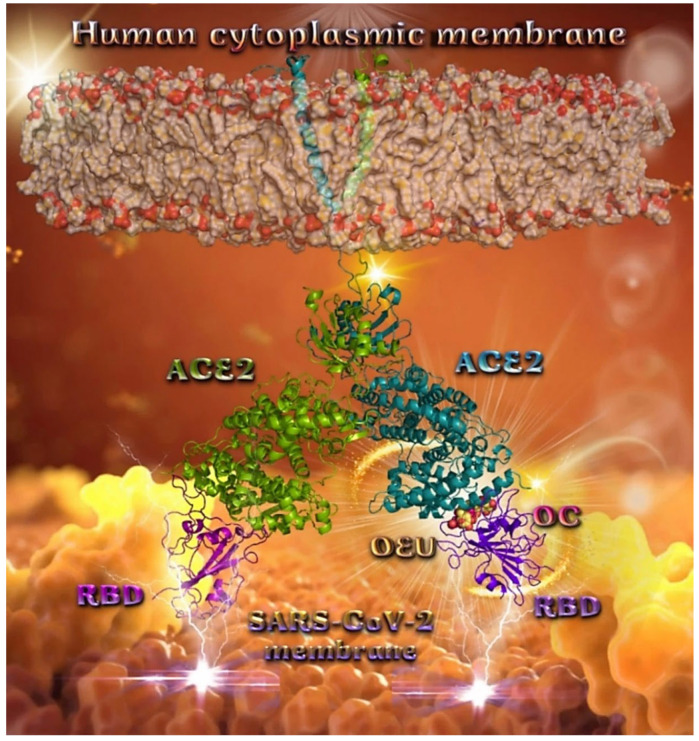
Figure 11.
Docking pose orientation of best-bound OEU and OC molecules on the crystal structure of wild-type (wt) SARS-CoV-2 full-length S protein’s RBD bound to the ACE2 receptor (from PDB ID: 6M17). The ACE2 receptor is depicted in its dimer structure with chains A and B with additional illustration of the transmembrane and extracellular domain. ACE2 receptor is shown to be embedded in a realistic membrane environment of lipid bilayer mimicking the human cytoplasmic membrane after molecular dynamics simulation. All structural models were downloaded from the Amaro lab (https://amarolab.ucsd.edu/covid19.php (accessed on 13 June 2021)). The protein complex is illustrated as cartoon colored by chain in deep purple for the RBD and deep teal for one monomer of the dimeric ACE2 receptor. OEU and OC molecules are rendered in sphere mode and colored according to atom type in yellow-orange and hot-pink C atoms, respectively. Both OEU and OC molecules are shown to be anchored in the interface between the RBD and ACE2 proteins. Color code used for lipid tails (surface representation): POPC, POPE, POPI, POPS, and cholesterol in wheat. P atoms of the lipid heads and cholesterol’s O3 atoms are highlighted in red. Molecular docking simulations were performed individually. Hydrogen atoms are omitted from both molecules for clarity. Heteroatom color code: O—red. The final structure was ray-traced and illustrated with the aid of PyMol Molecular Graphics Systems.