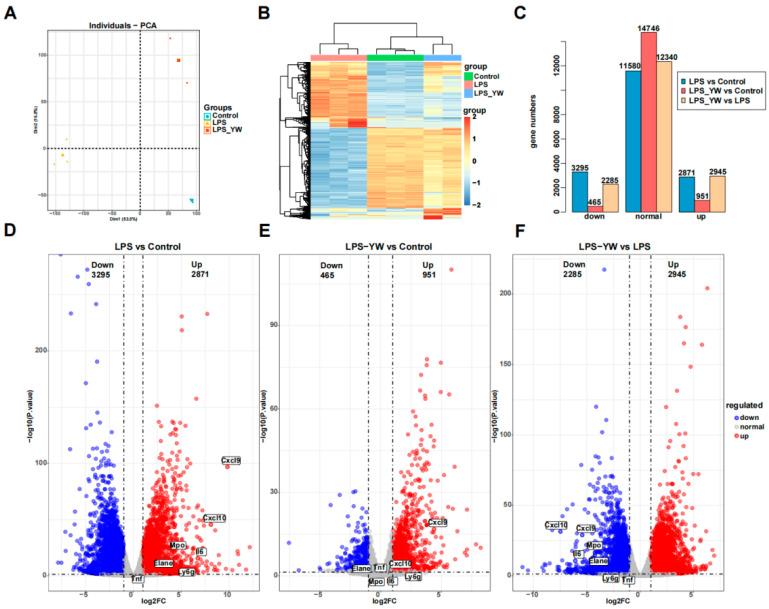
Figure 5.
RNA-seq data analysis of lung tissues of mice from different treatment groups. (A) Principal component analysis found that there was no overlap between the three groups (Control group, n = 3; LPS group, n = 3; LPS/YW3-56 dual-treatment group, n = 2). (B) Hierarchical clustering of the 1000 most variable genes also demonstrated clear separation of three group of samples. (C,D) Differential expression analysis identified significant numbers of differential expressed genes in LPS relative to control samples (total 6166 genes with 2871 upregulated and 3295 downregulated). (C,E) Less numbers of differential expressed genes (1416 genes) in LPS and YW3-56 dual treatment relative to control samples were found, of which 951 genes were upregulated and 465 genes were downregulated. (C,F) 5230 of differential expressed genes in LPS and YW3-56 dual treatment relative to LPS treatment samples were found, of which 2924 genes were upregulated and 2285 genes were downregulated. PCA, principal component analysis; log2FC, log2 fold change; LPS, lipopolysaccharides treatment gruoup, LPS_YW, LPS and YW3-56 dual-treatment group.