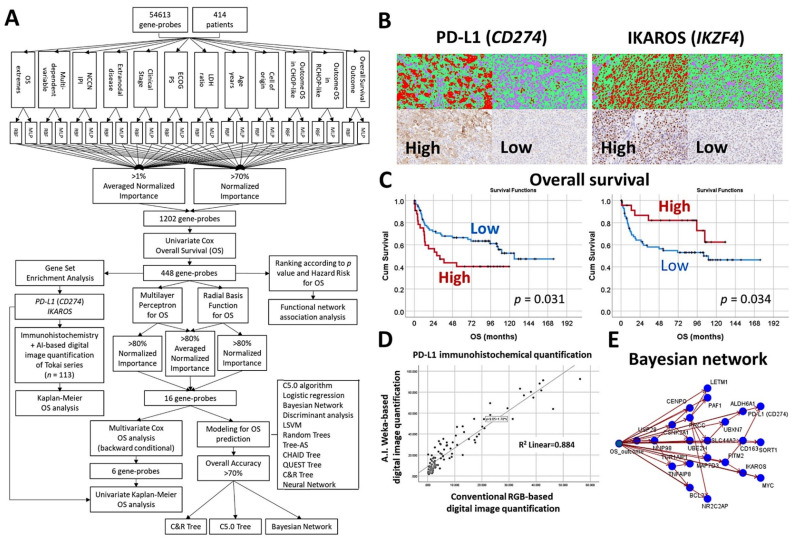
Figure 15.
An algorithm that included artificial neural networks and machine learning predicted the survival of diffuse large B-cell lymphoma, and highlighted PD-L1 and IKAROS as prognostic markers. (A) Algorithm: This algorithm is similar to that one of follicular lymphoma and mantle cell lymphoma. The basic structure analysis is an artificial neural network (multilayer perceptron). In this analysis, 54,613 gene probes were used as predictors for the overall survival, but also for other relevant clinicopathological variables. The basic neural network was composed of the input layer (predictors, 54,613 gene probes), a hidden layer (automatically computed), and an output layer (predicted variable; for example, the overall survival outcome as a dichotomic variable dead vs alive, or the cell of origin classification (GCB vs ABC), etc.). The dimensionality reduction included additional steps of machine learning, Cox regression, and GSEA. (B) Digital image quantification using AI-based strategy for PD-L1 (CD274) and IKAROS. (C) High protein expression of PD-L1 correlated with poor survival of the patients. Conversely, high IKAROS was associated with favorable survival. (D) AI-based quantification correlated well with conventional digital image quantification. Therefore, both techniques provide comparable results. (E) Modeling of the overall survival using a Bayesian network. The Bayesian network builds a probability model, a graphical model that shows variables (nodes) of the dataset, and the probabilistic (conditional) independences between them. The links of the network are called arcs and represent the relationship between the variables, but do not necessarily mean cause and effect. Original magnification: 200×. OS, overall survival; NCCN IPI, National Comprehensive Cancer Network International Prognostic Index; ECOG PS, Eastern Cooperative Oncology Group Performance Status; LDH, lactate dehydrogenase; R-CHOP, rituximab, cyclophosphamide, doxorubicin hydrochloride, vincristine, and prednisolone; AI, artificial intelligence.