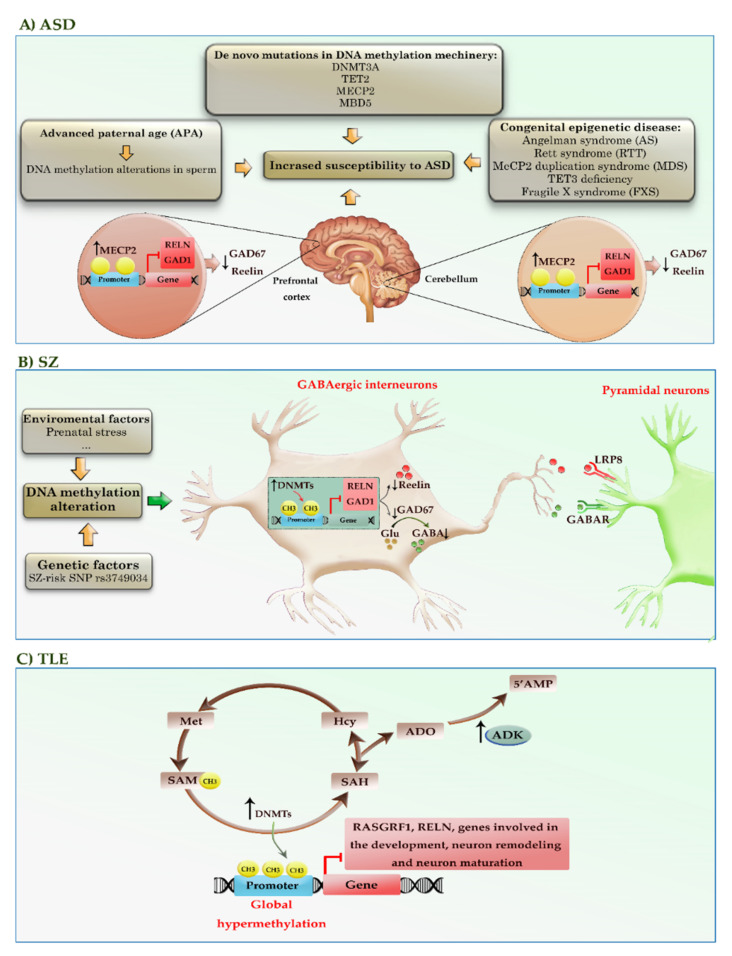
Figure 6.
A schematic representation of the DNA methylation alterations in the pathogenesis of neurodevelopmental and neuropsychiatric diseases, such as ASD, SZ, and TLE. (A) Advanced paternal age (APA), congenital epigenetic diseases, and de novo mutations in genes encoding DNA methylation machinery may increase the susceptibility of offspring to ASD. In addition, increased MECP2 interaction with RELN and GAD1 gene promoters triggers the reduction of Reelin and GAD67 expression in the brain regions of patients with ASD. (B) Environmental factors and certain genetic factors (e.g., rs3749034, an SZ-risk SNP) may cause aberrant methylation and associated silencing of genes, such as RELN and GAD1. In addition, the increased binding of MECP2 into the promoter of these genes can reduce the expression of Reelin and GAD67. Decreased GAD67 leads to reduced GABA release at synapses and compensatory regulation of postsynaptic GABAA receptors located in pyramidal neurons. (C) The alterations in the transmethylation pathway and adenosinergic signaling can lead to global hypermethylation of the genome in epileptogenic areas of the brain. Red colors highlighted hypermethylated genes. ADO: adenosine; AHCY: adenosylhomocysteinase; AMP: adenosine monophosphate; ADK: adenosine kinase; GAD1, 67: glutamic acid decarboxylase1, 67; GABA: gamma-aminobutyric acid; Hcy: homocysteine; RELN: Reelin; RASGRF1: Ras protein-specific guanine nucleotide releasing factor 1; SAM: S-adenosyl methionine; SAH: S-adenosyl homocysteine; SNP: single-nucleotide polymorphism; Met: methionine.