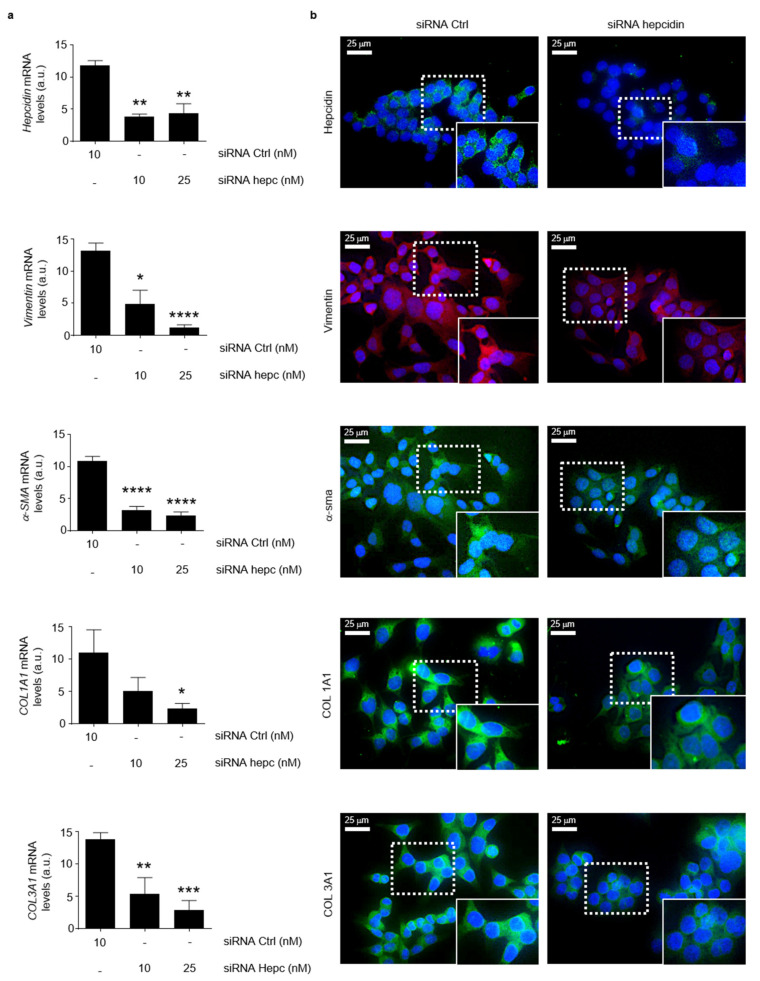
Figure 8.
Silencing of hepcidin in CRC cells is associated with a significant reduction of mesenchymal markers. (a) Hepcidin, Vimentin, α-SMA, COL1A1 and COL1A3 RNA transcripts were evaluated in HCT-116 cells transfected for 24 h with either a control siRNA (10 nM) or hepcidin siRNA Hepcidin (10 and 25 nM) by real-time PCR, and values were normalized to β-actin RNA. Values are expressed in arbitrary units (a.u.) and indicate mean ± SD of 4 experiments; Hepcidin: control siRNA versus hepcidin siRNA 10 nM, ** p < 0.01; control siRNA versus hepcidin siRNA 20 nM, ** p < 0.01; Vimentin: control siRNA versus hepcidin siRNA 10 nM, * p < 0.05; control siRNA versus hepcidin siRNA 20 nM, **** p < 0.0001; α-SMA: control siRNA versus hepcidin siRNA 10 nM, **** p < 0.0001; control siRNA versus hepcidin siRNA 20 nM, **** p < 0.0001; COL1A1: control siRNA versus hepcidin siRNA 10 nM, ns; control siRNA versus hepcidin siRNA 20 nM, * p < 0.05; COL1A3: control siRNA versus hepcidin siRNA 10 nM, ** p < 0.01; control siRNA versus hepcidin siRNA 20 nM, *** p < 0.001. (b) Representative images of immunofluorescence staining for hepcidin (green), Vimentin (red), α-SMA (red), COL1A1 (green) and COL1A3 (green) in HCT-1116 cells transfected with either a control siRNA (10 nM) or hepcidin siRNA Hepcidin (25 nM) for 24 h; nuclei are stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue).