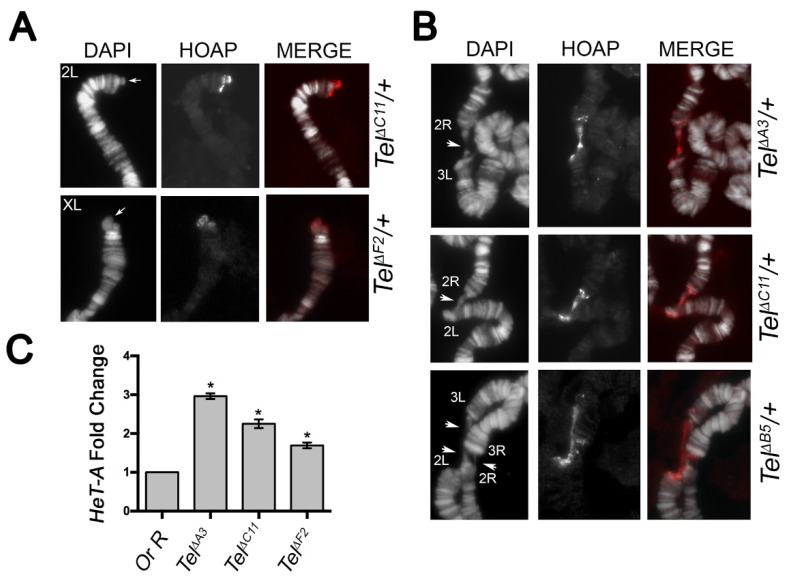
Figure 7.
Telomere elongation in the CRISPR/Cas 9-induced Tel deletion alleles. (A) anti-HOAP immunostaining on 2L and XL chromosome tips from Tel∆F2/+ and Tel∆C11/+ hemyzygotes, respectively, showing protruding telomeric DNA only on the Tel mutant chromosomes (arrows). Note that, as expected, Tel mutant elongated telomeres do not impair HOAP localization. (B) Examples of telomere fusions involving either two or four chromosome tips (arrowheads) from different Tel∆ hemizygotes. (C) qPCR analysis on third instar larval DNA from two representative Tel∆ deletion alleles showing a robust increase in Het-A copy number compared to control (OR-R). Note that Tel∆B5 bears a deletion that uncovers TGT, while Tel∆C10 does not include TGT. * (p < 0.05).