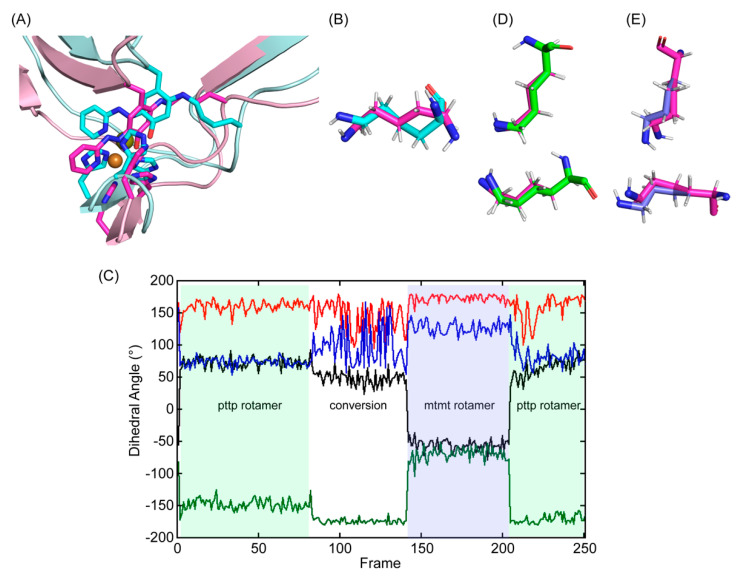
Figure 6.
Molecular modeling and energy minimization of 2HP–inhibited Δ1–2SRCR–LOXL2 in comparison to the structure after MD–simulation. (A) The initial 3D–model (in cyan) was generated by replacing Tyr689 with DPQ–2HP and crosslinking C2 with the ϵ–amino side chain of Lys653. The single bond between ϵ–nitrogen and δ–carbon of Lys653 and the aromatic ring of Tyr698 (LTQ–2HP) was not on the same plane (dihedral angle = 15°) and the –(CH2)4–NH– side chain of Lys653 was in a higher energy conformation. After manual rearrangement of the hexapeptide (His652–Lys653–Ala654–Ser655–Phe656–Cys657) and subsequent MD–simulation, the –(CH2)4–NH– side chain of Lys653 was in a more relaxed conformation (in magenta and light pink). (B) An overlaid view of Lys653 in the initial 3D–model (in cyan) and in the final structure after MD–simulation (in magenta) (C) Dihedral angles of Lys653 during the 10 ns MD–simulation (40 ps/frame): Χ1 (dihedral angle between N–Cα–Cβ–Cγ) in black; X2 (dihedral angle between Cα–Cβ–Cγ–Cδ) in red; X3 (dihedral angle between Cβ–Cγ–Cδ–Cϵ) in green; X4 (dihedral angle between Cγ–Cδ–Cϵ–Nζ) in blue. We observed pttp and mtmt rotamers among the ideal rotamers reported for Lys653 in the penultimate rotamer library: t stands for the side chain dihedral angles of 180°, p stands for the side chain dihedral angles of 65°, and m stands for the side chain dihedral angles of – 65° [31]. (D) An overlaid view of Lys653 after MD–simulation (in magenta) with the pttp rotamer (in green) (E) An overlaid view of Lys653 after MD–simulation (in magenta) with the mtmt rotamer (in slate).