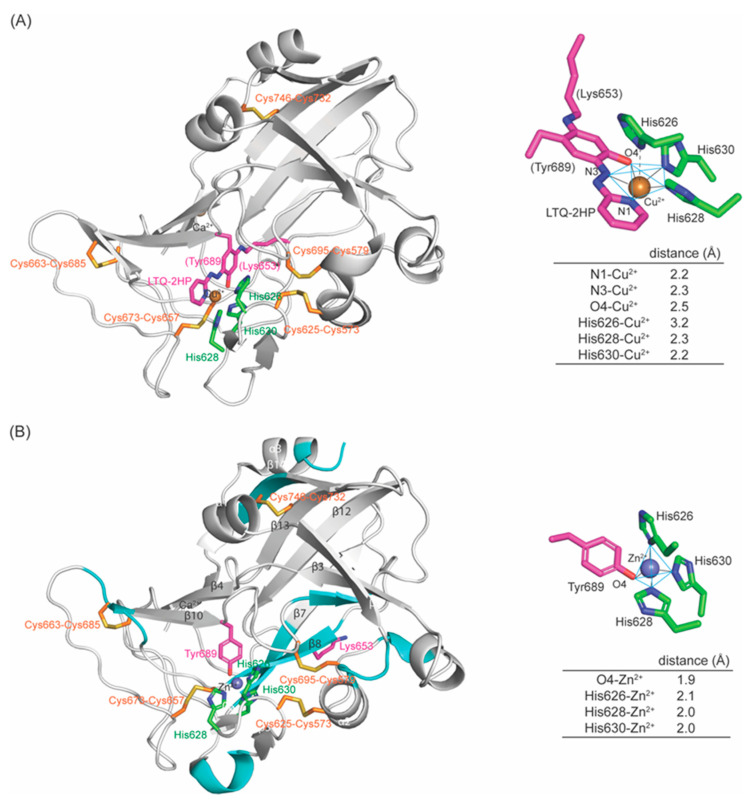
Figure 10.
Comparison of amine oxidase domains and metal–binding sites of the 3D–modeled 2HP–inhibited LOXL2. (A) and the crystal structure of Zn2+–bound precursor (PDB:5ZE3) (B). Overall structures are very similar (RMSD = 1.347). In 3D–modeled structure (A), the cyan–colored secondary structure features in the precursor (B) are missing (having become loops). In the 2HP–ihibited LOXL2 (A), the Cu2+–coordination environment is a distorted square pyramidal with O4–Cu2+ in a Jahn–Teller axis and N1 and N3 of LTQ–2HP and His628 and His630 comprising the square bottom. In the precursor structure (B), Zn2+ is in tetrahedral coordination geometry having O4, His626, His628 and His630 at each tip of the tetrahedron. The cysteine–paring pattern of the five disulfide bonds are totally conserved in both structures.