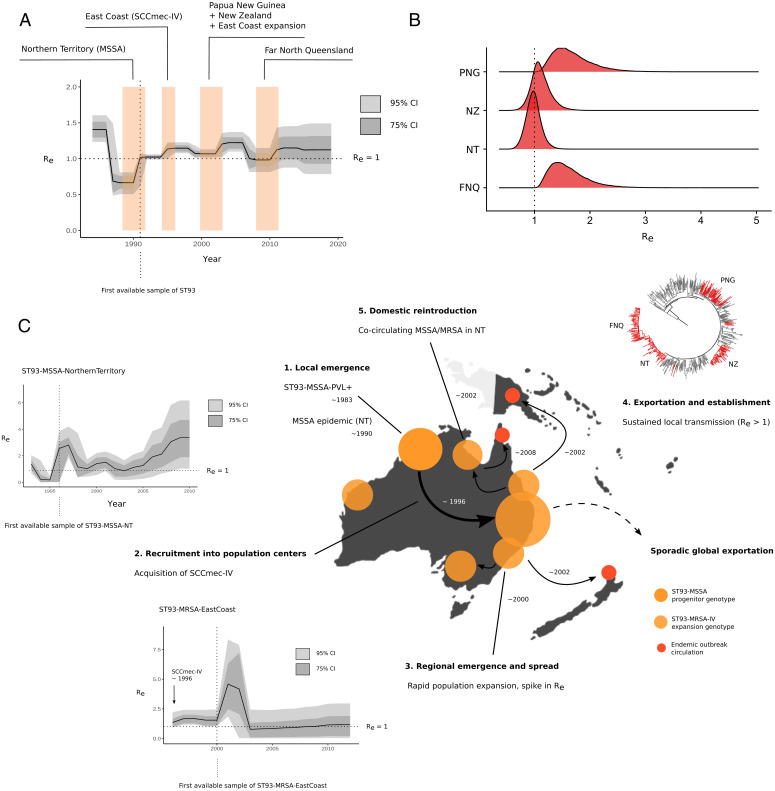
Fig. 2.
Phylodynamic signatures and parameter estimates for the Australian lineage (ST93) (A) Changes in Re over time, showing the 95% and 75% credible intervals (CIs) of the MRCA of clade-divergence events of ST93-MSSA and ST93-MRSA (colored blocks) using the birth–death skyline model. (B) Re posterior density distributions for introductions in PNG, FNQ, NZ, and reintroduction of the MRSA genotype to the NT. (C) Events in the emergence and regional dissemination of the QLD clone, with ML phylogenies and branch colors indicating major subclades and divergence events in the emergence of ST93. Vertical lines in skyline plots indicate the year of the first sample from the lineage or clade, horizontal lines indicate the epidemic threshold of Re = 1 (step 1). Local emergence of ST93-MSS in remote Indigenous communities of north Western Australia. Some sporadic transmission to QLD occurred in this clade. ST93-MSSA continues to circulate in the NT (NT). Step 2: Re > 1 estimates indicate that strains continue to spread (Inset Plot). Step 3: When the lineage acquired SCCmec-IV, it spread to East Australian coastal states (QLD, VIC, NSW) coinciding with population growth and increase in transmission with a spike at initial recruitment (Inset Plot). Step 4: ST93-MRSA-IV continues to spread in the eastern coastal states (QLD, NSW, VIC). Step 5: From the globally connected East Coast population centers, ST93-MRA-IV spread overseas (particularly to the United Kingdom) but also established sustained transmission in remote FNQ and regionally in the highlands of PNG. The FNQ outbreak is derived from an ST93-MRA-IV clade in the NT cocirculating with the ongoing ST772-MSSA epidemic.