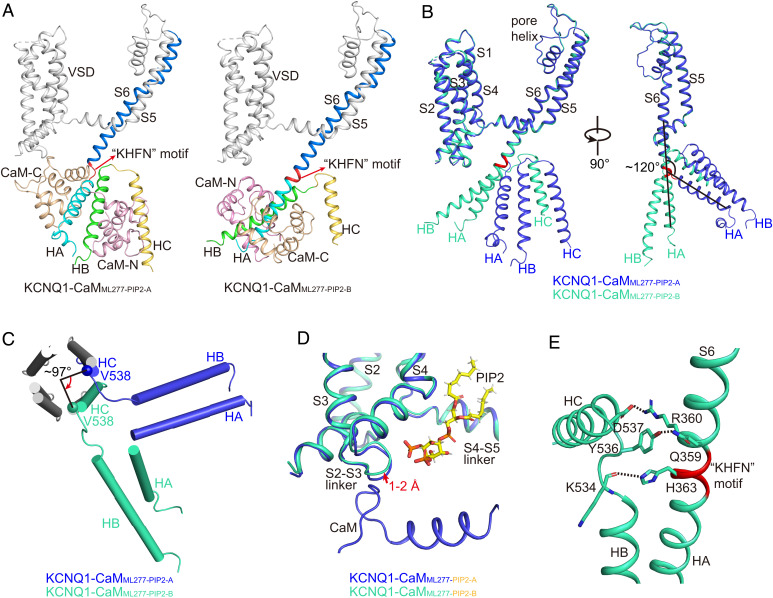
Fig. 7.
Structural comparisons of KCNQ1-CaMML277-PIP2-A and KCNQ1-CaMML277-PIP2-B. (A) Different structure arrangements of CTD and CaM in KCNQ1-CaMML277-PIP2-A and KCNQ1-CaMML277-PIP2-B. S6, S6-HA linker, HA, HB, and HC helices of KCNQ1 are colored in marine, red, cyan, green, and yellow, respectively. The N-lobe and C-lobe of CaM are shown in wheat and pink, respectively. (B) Structural comparison of one KCNQ1 subunit in KCNQ1-CaMML277-PIP2-A (blue) and KCNQ1-CaMML277-PIP2-B (green) in two side views. The S6-HA linker ‘‘KHFN’’ motif that undergoes structural rearrangement from a loop to a helix is colored in red. (C) Structural comparison of the HA, HB, and HC helices in KCNQ1-CaMML277-PIP2-A (blue) and KCNQ1-CaMML277-PIP2-B (green) in the top view with TMD and HA/HB from the other three subunits omitted for clarity. The N-terminal residue Val538 of HC rotates by ∼97° from KCNQ1-CaMML277-PIP2-A to KCNQ1-CaMML277-PIP2-B. (D) The binding of PIP2 induces a 1–2 Å movement of the S2-S3 linker in KCNQ1-CaMML277-PIP2-B (green) in comparison with that in KCNQ1-CaMML277-PIP2-A (blue). The PIP2 in KCNQ1-CaMML277-PIP2-A is not shown for clarity. (E) Interactions between S6 and HB-HC linker. The dashed lines indicate the salt bridge Arg360-Asp537 and the hydrogen bonds Gln359-Tyr536 and His363-Lys534.