Abstract
Fructooligosaccharides (FOS) promote the proliferation of Bifidobacterium, especially Bifidobacterium pseudolongum in C57BL/6J mice. However, the response of intestinal microbes to FOS is influenced by host genotypes. Therefore, we compared the intestinal microbiota of four commonly used mice before and after FOS intervention, including C57BL/6J, BALB/c, Institute Cancer Research (ICR), and Kunming (KM) mice. The intestinal microbiota of the four genotypes exhibited similarities in composition but differences in relative abundance. Bifidobacterium was significantly increased to different degrees in the four genotypes of mice after FOS intervention, and Akkermansia and Bacteroides were also significantly increased in BALB/c and KM mice. Lactobacillus and Alistipes levels were unchanged or decreased. Within the genus Bifidobacterium, B. pseudolongum was the dominant species in the four genotypes of mice and proliferated significantly after FOS intervention, with dramatic proliferation in C57BL/6J mice (9.49%). Furthermore, eight strains of B. pseudolongum were screened from the feces of mice with four genotypes, and there was a great difference in the ability and manner of utilizing FOS among the strains. The strains from C57BL/6J mice exhibited the strongest utilization of 1-kestose (GF2), whereas other strains could utilize both GF2 and nistose (GF3) weakly. The gut microbial analysis of mice with different genotypes complemented our previous studies. The results provided the background strains of the different mouse genotypes and suggested a correlation between the utilization ability and the response of the strains to FOS. Further studies on the utilization ability of strains and competition in the intestine will contribute to the understanding of the mechanisms of the intestinal microbial response to diet.
Keywords: FOS, Intestinal microbiota, Genotypes, B. pseudolongum
Graphical abstract
FOS significantly increased the relative abundance of intestinal B. pseudolongum in four genotypes of mice, with the most significant effect on C57BL/6J mice. The ability and manner of FOS utilization by B. pseudolongum strains from different genotypes of mice varied greatly, with strains derived from C57BL/6J mice being more capable of utilizing FOS.
Highlights
-
•
The intestinal microbiota is similar in composition for mice of different genotypes.
-
•
B. pseudolongum predominates in bifidobacteria in mice of different genotypes.
-
•
The relative abundance of B. pseudolongum increases after FOS intake.
-
•
B. pseudolongum strains show different abilities in utilizing FOS.
1. Introduction
In recent years, the composition of intestinal microbiota has received increasing attention as a factor affecting host health. The intestinal microbiota of mice has been extensively studied to reveal the relationship between the intestinal microflora and host health. Mouse models are crucial components of gut microbiome research. However, no specific mouse model is recommended for dietary interventions, and in-house or commercially available strains are commonly chosen (Beck et al., 2000; Hildebrand et al., 2013; Hugenholtz and de Vos, 2018). This may not be desirable because the genetic backgrounds, experimental settings, individual differences, and various other factors of the mouse microbiota complicate cross-study comparisons and a global understanding of mouse microbiome patterns (Hildebrand et al., 2013).
Previous studies have compared the effects of genotype (Goodrich et al., 2014), age, diet, and other factors on the composition of the intestinal microbiota. The results indicated that diet had a significant and rapid effect on the composition of intestinal microbiota, whereas age showed a weak but lasting impact on gut microbiota (Org et al., 2015; Parks et al., 2013). Furthermore, sex was found to have no or very limited impact on intestinal microbiota (Kovacs et al., 2011). The shaping effects of diet and age on the gut microbiota vary with different genotypes (Xu et al., 2022). Therefore, genetic differences among mice with different genotypes may be an important factor in determining the gut microbial diversity and overall structure (Carmody et al., 2015; Spor et al., 2011).
The gut microbiota fulfills a variety of functions in the host, including degradation of dietary carbohydrates. The existence of indigestible carbohydrates in the diet, especially oligosaccharides, has been widely studied to determine the impact of intestinal microflora; among them, Fructooligosaccharides (FOS) have been reported to increase the relative abundance of Bifidobacterium in BALB/c and C57BL/6J mice (Wang et al., 2017). Our previous studies further confirmed that FOS promote the proliferation of B. pseudolongum in C57BL/6J mice (Mao et al., 2018). However, the host genotype plays an important role in determining host-microbe interactions (Bonder et al., 2016; Khachatryan et al., 2008), which might lead to different profiles of intestinal microbiota after FOS ingestion. Therefore, it is necessary to investigate the intestinal microbial composition and the effect of FOS on the composition of the intestinal microbiota in mice with different genotypes. Meanwhile, the detection of the response of B. pseudolongum in mice with different genotypes to FOS will complement our previous studies and would also contribute to the understanding of the response of intestinal microorganisms present in mice with diverse genetic backgrounds to FOS.
In this study, Illumina MiSeq sequencing and bioinformatics analysis were used to analyze the intestinal microbiota of the commonly used experimental mice, Kunming (KM), Institute Cancer Research (ICR), BALB/c, and C57BL/6J mice before and after FOS administration. By analyzing the intestinal microbiota of multiple genotypes of mice, we revealed the diversity of the intestinal microflora of commonly used mice. The results were compared to provide a reference for the selection of murine Furthermore, B. pseudolongum strains were isolated from the four genotypes of mice to compare the ability of B. pseudolongum from mice of different genotypes to utilize FOS models in intestinal microbial studies.. The effects of FOS on the composition of intestinal microflora in different genotypes of mice and the ability of B. pseudolongum strains to utilize FOS were studied to understand the mechanism of interaction between prebiotics and intestinal microflora.
2. Material and methods
2.1. Chemicals and reagents
FOS (GFn type, high-performance liquid chromatography purity: 95.93%) were purchased from Bao Lingbao Biotechnology (Shandong, China). FOS are a mixture of oligosaccharides consisting of one, two or three fructose units (F) bound to the β-2, 1 position of sucrose (GF) (Sangeetha et al., 2005). The main components were 1-kestose (GF2: 60.60%), nistose (GF3: 32.45%), and 1F-fructofuranosylnystose (GF4: 2.88%). The reagents used for bacterial culture were obtained from Sinopharm Chemical Reagents (Shanghai, China), including proteose peptone, papaic digest of soybean meal, yeast extract, beef extract, digestive serum, KH2PO4, NaCl, and L-cysteine hydrochloride (Gu et al., 2022b).
2.2. Animal experiment design
Eight-week-old male C57BL/6J, BALB/c, ICR, and KM mice were purchased from Charles River (Shanghai, China). Sixteen mice of each genotype were randomly assigned to FOS and control groups (n = 8/group), and four mice were kept in a cage. All mice were fed in an environment with a 12-h light/dark cycle under constant temperature (22 °C) and allowed access to water and food ad libitum. All experiments in this study were approved by the Ethics Committee of Jiangnan University (JN. No20210515m0800710[121]) and were performed in accordance with the EU guidelines for experimental animals (Directive, 2010/63/EU).
The mice in the FOS and control groups were orally administered FOS (7.5 g/kg/day) and sterile water, respectively, for 4 weeks after a week of adaption. At the end of the experiment, the colon and the cecum were collected and weighed. The microbiota composition of the feces and cecal contents were analyzed using the MiSeq platform (Illumina, San Diego, CA, USA). All samples were stored at −80 °C for further assays.
2.3. Analysis of SCFAs level in feces of mice
SCFAs were analyzed by Gas Chromatographic Mass Spectrometer (GC-MS), as described previously, with slight modifications (Mao et al., 2016). In brief, lyophilized feces were acidified with 20 μL of 10% sulfuric acid. Then, 0.8 mL of ether was added, and the mixture was mixed before centrifugation (14,000×g for 5 min). The organic phase was separated into 0.25 g of anhydrous sodium sulfate to remove the excess water. The centrifuged supernatant was transferred to a vial to determine the concentrations of acetic, propionic, butyric, isobutyric, valeric, and isovaleric acids.
2.4. DNA extraction, polymerase chain reaction (PCR) amplification, sequencing, and bioinformatics analysis
Fecal and cecal contents from mice were collected to assess changes in the composition of the intestinal microbiota. Bacterial genomes in mouse feces and cecal contents were extracted using the FastDNA Spin Kit for Feces (MP Biomedicals, LLC, Irvine, CA, USA) according to the manufacturer's instructions. The V3–V4 region of the 16S rDNA and groEL gene were amplified. Quantification, sequencing, and bioinformatics analyses were performed as previously described (Mao et al., 2018).
2.5. Isolation and identification of B. pseudolongum in mouse feces
Fecal samples of mice with different genotypes were collected and suspended in 9 g/L NaCl solution. The suitable diluent was plated on the Gifu Anaerobic Medium (GAM) with FOS as the sole carbon source. The selected strains were purified and identified using 16S rRNA sequencing. The specific experimental steps of the screening were performed as we previously described (Gu et al., 2022b).
2.6. Differences in FOS utilization during the growth of B. pseudolongum
Isolated B. pseudolongum strains were inoculated in GAM medium under anaerobic conditions. The OD600 was measured and the supernatant was collected every 1 or 2 h. High-performance liquid chromatography (HPLC) analysis of FOS in the supernatants of bacterial cultures was performed to compare differences in FOS utilization among the B. pseudolongum strains. The contents of GF2, GF3 and GF4 were detected using the XBridge ® BEH amino column (Waters, USA, length, 250 mm; internal diameter [i.d.], 4.6 μm) at a column temperature of 35 °C, with a flow phase of acetonitrile-water (75:25) at a flow rate of 1 mL/min. The content was determined with reference to an external standard (Gu et al., 2022b).
2.7. Statistical analysis
Statistical analyses were performed using Origin 2018 and SPSS 16, and the results are presented as mean ± standard error (SEM). The differences among groups were judged by ANOVA and the differences between two groups were judged by the t-test. Statistically significant differences are indicated with asterisks as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3. Results
3.1. Body weight, dietary intake, and serum biochemical indicators
The weight and dietary intake of mice were recorded daily during the intervention period (Fig. 1). The weight gain in the FOS group was slightly lower than that in the control group. No significant difference in body weight was observed before and after FOS administration among the four genotypes of mice (p > 0.05). The body weights of BALB/c and C57BL/6J mice were 24.06 ± 0.98 g and 24.13 ± 1.59 g, and that of ICR mice was 39.57 ± 2.25 g. The weight gain of KM mice reached 5.88 ± 2.07 g and 6.79 ± 3.79 g, respectively in FOS and control groups. The average daily food intake of the mice also showed no significant difference between the two groups during the intervention period (p > 0.05), ranging from 2.07 g to 4.36 g in the BALB/C and C57BL/6J groups. The dietary intake of ICR mice fluctuated from 3.0 g to 6.69 g, and that of KM mice was from 6.01 g to 14.38 g. Besides, the serum biochemical markers, including serum glucose (Glu), total cholesterol (TC), triglyceride (TG), low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) levels also showed no significant differences between the FOS and control groups (p > 0.05, Table 1).
Fig. 1.
The body weight (A) and daily diet intake (B) of mice.
Table 1.
Serum biochemical markers in mice.
| Group | Glu (mmol/L) | HDL-C (mmol/L) | LDL-C (mmol/L) | HDL-C/LDL-C | TC (mmol/L) | TG (mmol/L) |
|---|---|---|---|---|---|---|
| C57-Control | 1.46 ± 0.68 | 0.76 ± 0.11 | 0.14 ± 0.02 | 5.38 ± 0.65 | 1.04 ± 0.15 | 1.00 ± 0.13 |
| C57-FOS | 0.82 ± 0.35 | 0.71 ± 0.09 | 0.16 ± 0.03 | 4.46 ± 0.67 | 1.03 ± 0.11 | 1.02 ± 0.19 |
| Ba-Control | 0.83 ± 0.35 | 0.99 ± 0.05 | 0.15 ± 0.01 | 6.50 ± 0.40 | 1.27 ± 0.07 | 1.39 ± 0.11 |
| Ba -FOS | 1.15 ± 0.49 | 1.07 ± 0.09 | 0.17 ± 0.02 | 6.49 ± 1.12 | 1.43 ± 0.14 | 1.79 ± 0.27 |
| KM-Control | 0.93 ± 0.28 | 0.91 ± 0.15 | 0.16 ± 0.06 | 5.93 ± 1.45 | 1.23 ± 0.25 | 1.14 ± 0.32 |
| KM-FOS | 0.87 ± 0.20 | 0.88 ± 0.16 | 0.17 ± 0.06 | 5.69 ± 0.97 | 1.21 ± 0.26 | 1.18 ± 0.28 |
| ICR-Control | 0.72 ± 0.61 | 1.29 ± 0.27 | 0.30 ± 0.09 | 4.56 ± 0.96 | 1.84 ± 0.43 | 1.22 ± 0.35 |
| ICR-FOS | 0.55 ± 0.42 | 1.16 ± 0.25 | 0.22 ± 0.05 | 4.91 ± 2.54 | 1.53 ± 0.38 | 1.07 ± 0.41 |
Notes: The serum biochemical markers between the Control and FOS groups of the four mice strains showed no significant differences (t-test, p > 0.05).
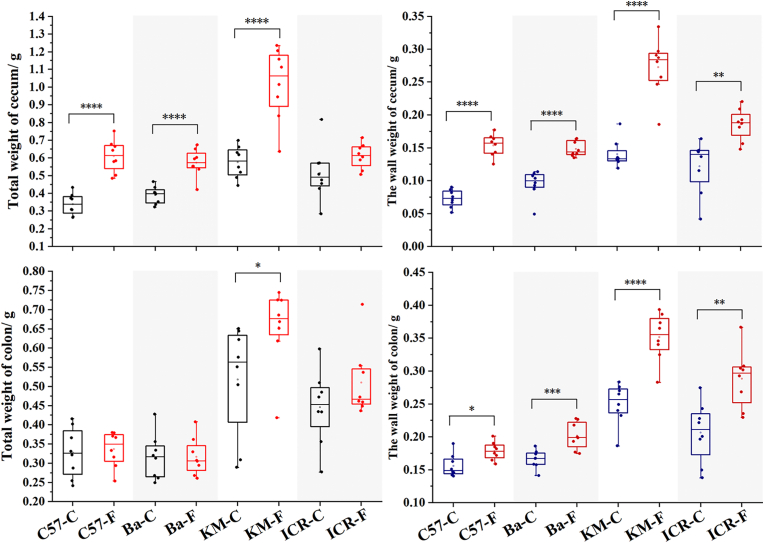
3.2. Total and wall weight of the cecum and colon
The cecum and colon were collected and weighed to obtain the total weight. The wall weights of the cecum and colon were measured after the removal of the contents (Fig. 2). FOS significantly increased the total and wall weights of the cecum in mice, in addition to the total weight of the cecum in ICR mice. The total cecal weight of C57BL/6J mice in the FOS group was 1.80 times heavier than that in the control group, which was the most obvious increase among the four genotypes. The cecum of KM mice was the heaviest (0.58 ± 0.09 g) and increased to 1.02 ± 0.20 g after FOS administration. The cecal wall weight of C57BL/6J mice in the control and FOS groups were 0.07 ± 0.01 g and 0.15 ± 0.02 g, respectively. FOS had the greatest effect on the cecal wall weight in C57BL/6J mice. FOS also significantly increased the colonic wall weight of mice and had the most significant effect on the total colonic weight and colonic wall weight of KM mice. The colon wall weight of KM mice was 0.25 ± 0.03 g in the control group and 0.35 ± 0.04 g in FOS group.
Fig. 2.
Total weight and wall weight of the cecum and colon.
3.3. Short-chain fatty acids (SCFAs) determination in mice feces
FOS can be fermented by intestinal microbiota to produce short-chain fatty acids. After FOS intervention, the contents of acetic acid and propionic acid in mice feces increased to varying degrees. The greatest increase in acetic acid was observed in the feces of C57BL/6J mice and the greatest increase in propionic acid was observed in the feces of ICR mice (Table 2). The content of butyric acid was increased in the feces of C57BL/6J and ICR mice, while it was significantly reduced in the feces of KM mice. There is no significant difference in the content of total acid in the feces, and the highest content of total acid reached 286.70 ± 75.86 nmol/g in the feces of ICR mice.
Table 2.
Concentration of short-chain fatty acids (SCFAs) in feces of mice in FOS and control groups.
| SCFAs, nmol/g | Acetic acid | Propionic acid | Butyric acid | Isobutyric acid | Valeric acid | Isovaleric acid | Total acid |
|---|---|---|---|---|---|---|---|
| C57-Control | 110.01 ± 37.09 | 42.33 ± 13.18 | 22.42 ± 7.08 | 12.66 ± 4.15 | 13.55 ± 4.35 | 11.27 ± 4.02 | 212.24 ± 68.52 |
| C57-FOS | 132.26 ± 23.50 | 45.83 ± 6.26 | 26.20 ± 4.25 | 11.21 ± 3.56 | 11.87 ± 3.82 | 9.65 ± 2.79 | 237.01 ± 36.85 |
| Ba-Control | 149.06 ± 25.96 | 41.91 ± 5.81 | 36.91 ± 12.13 | 10.76 ± 1.94 | 11.59 ± 2.07 | 8.85 ± 1.74 | 259.08 ± 47.44 |
| Ba-FOS | 150.17 ± 9.71 | 43.10 ± 5.37 | 25.89 ± 4.84 | 10.58 ± 1.47 | 11.21 ± 1.65 | 8.79 ± 1.23 | 249.74 ± 19.85 |
| ICR-Control | 139.96 ± 55.80 | 45.24 ± 14.62 | 37.14 ± 18.42 | 12.59 ± 1.92 | 13.48 ± 1.84 | 10.18 ± 1.34 | 258.59 ± 92.74 |
| ICR-FOS | 155.69 ± 42.39 | 56.03 ± 15.67 | 38.00 ± 16.26 | 12.75 ± 3.04 | 13.54 ± 3.52 | 10.68 ± 2.32 | 286.70 ± 75.86 |
| KM-Control | 144.61 ± 24.24 | 41.74 ± 11.65 | 35.20 ± 14.50 | 11.28 ± 4.15 | 12.22 ± 4.54 | 9.75 ± 3.84 | 262.92 ± 59.02 |
| KM-FOS | 157.22 ± 14.95 | 48.97 ± 9.22 | 25.60 ± 8.00 | 9.43 ± 1.51 | 9.72 ± 1.64 | 7.64 ± 1.17 | 258.58 ± 35.04 |
Notes: The concentration of SCFAs in feces of mice in FOS and control groups showed no significant differences (t-test, p > 0.05).
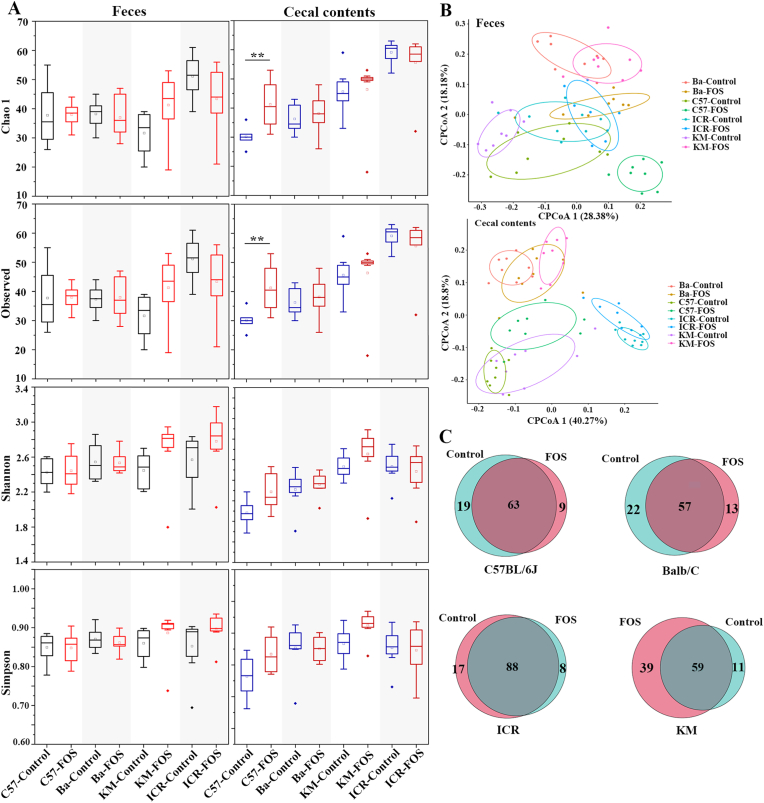
3.4. Diversity of intestinal microflora in different mice genotypes
α-Diversity was evaluated using the Chao1, Observed, Shannon, and Simpson indices (Fig. 3A). After FOS administration, the α-diversity of the feces showed no statistically significant differences compared to that of the control groups. Chao1 and Observed indices of cecal contents were significantly enhanced in C57BL/6J mice, indicating that FOS increased the microbial richness of cecal contents in C57BL/6J mice (p < 0.01). β-diversity was reflected by principal coordinates analysis (PCoA) of the weighted UniFrac distance, which provides a measurement of distance or dissimilarity between each sample. The distributions of the examined fecal and cecal content samples of the FOS and control groups are plotted in Fig. 3B. The intestinal microflora of BALB/c mice differed significantly from those of other mice. A distinct clustering pattern between the control and FOS groups was explicitly observed. The characteristics and common taxa of the FOS and control groups are shown in a Venn diagram (Fig. 3C). These results indicated that the structure of the intestinal microflora of the four genotypes of mice was significantly different, and FOS administration affected the structure of the intestinal microbiota.
Fig. 3.
α diversity (A), β diversity (B) and Veen diagram analysis (C) of intestinal microbiota of mice with different genotypes.
3.5. Effect of FOS on the cecal and fecal microbiota of mice
The intestinal microbiota of the mice was investigated using 16S rRNA sequencing. A total of 8,396,395 V3–V4 sequences were generated, and 151 genera belonging to 10 phyla were identified. At the phylum level, Firmicutes and Bacteroidetes were the most prevalent phyla, accounting for over 93.58%, and the proportions of these phyla varied a lot among the four genotypes of mice (Fig. 4A). Firmicutes-to-Bacteroidetes ratio (0.91) was the lowest in C57BL/6J mice. Verrucomicrobia accounted for 3.58% in C57BL/6J mice and Deferribacteres accounted for 4.07% and 1.57% in KM and ICR mice, respectively. After FOS administration, the relative abundance of Bacteroidetes and Actinobacteria significantly increased, whereas that of Firmicutes decreased in the four genotypes. The Firmicutes-to-Bacteroidetes ratio showed a significant decrease (p < 0.05). The relative abundance of Actinobacteria was the most significantly increased in C57BL/6J mice (p < 0.0001), from 0.33% to 13.94%. The relative abundances of Verrucomicrobia and Cyanobacteria were higher in BALB/c mice. And in ICR mice, the relative abundances of Proteobacteria (5.76%) and Verrucomicrobia (4.44%) also increased. These results indicated that there were some similarities, but also differences, in the intestinal microbiota of the four genotypes of mice. In addition, significant differences in the response to FOS at the phylum level were observed among the genotypes.
Fig. 4.
Effect of FOS on the cecal and fecal microbiota of mice. (A) the relative abundance of fecal microbiota in mice of different genotypes at the phylum level. (B) the composition of fecal and cecal microbiota at the genus level. (C) the key fecal microbiota in mice of different genotypes at the genus level after FOS administration.
The distinct variation in the intestinal microbiota at the genus level is presented in Fig. 4B. Forty-one out of the total genera comprised >97.29% of the intestinal microbiota. Before the intervention period, the genera belonging to Muribaculaceae family were the dominant taxa in the feces of ICR, KM, C57BL/6J, and BALB/c mice (43.33% and 42.14% in control and FOS groups). Besides, Alistipes and Lactobacillus accounted for more than 10% in the feces of ICR and C57BL/6J mice. Lachnospiraceae and Alloprevotella accounted for a large proportion in these samples. However, the cecal contents showed a different composition of microbiota than the feces. The relative abundance of Lachnospiraceae, Desulfovibrio, Roseburia, and Lachnoclostridium was higher than that in the feces, while there was a decrease in Lactobacillus and Muribaculaceae.
Despite the similar composition of intestinal microflora in the four genotypes of mice, the response of the microbiota to FOS was diverse in the four genotypes of mice. After FOS intervention, the composition of intestinal microbiota changed independently in the feces and cecal contents of the four genotypes of mice, except for the growth of Bifidobacterium in all genotypes of mice (Fig. 4C). Eleven genera showed significant variation in the feces of C57BL/6J mice. The relative abundance of Bifidobacterium increased sharply from 0.23% to 10.46% in the feces of C57BL/6J mice, and Coriobacteriaceae UCG-002, Prevotellaceae UCG-001, and Ruminococcus also increased. However, the relative abundance of Akkermansia (from 0.09% to 8.02%) increased more than that of Bifidobacterium (from 0.00% to 1.87%) in the feces of BALB/c mice, whereas the relative abundance of Lachnospiraceae and Lactobacillus showed an obvious decrease. In the feces of ICR mice, the relative abundance of Bifidobacterium, Akkermansia, and Muribaculaceae increased significantly while that of Alistipes decreased. FOS also had a significant effect on the fecal microbiota of KM mice. The relative abundances of Muribaculaceae and Bacteroides showed a maximum increase, whereas that of Bifidobacterium increased to 2.46%. The relative abundance of Lactobacillus and Alistipes decreased.
The microbial composition of the cecal contents was not consistent with that of feces before and after FOS administration at the genus level. FOS significantly promoted the proliferation of Bifidobacterium and decreased that of Lactobacillus and Alistipes in the cecal contents of mice. The relative abundance of Bifidobacterium in C57BL/6J mice increased from 1.09% to 12.23% and reached 0.76%, 2.42%, and 3.76% in the cecal contents of BALB/c, ICR, and KM mice, respectively.
3.6. Effect of FOS on the composition of bifidobacterium in mice
Furthermore, we explored the changes in the composition of bifidobacteria at the species level. Thirteen bifidobacteria species were detected among the samples from the four genotypes of mice using groEL sequencing. As shown in Fig. 5, B. pseudolongum, B. longum, B. breve and B. pseudocatenulatum were the predominant members of the Bifidobacterium in the feces and cecal contents. After FOS intervention, B. pseudolongum proliferated significantly and became the most abundant species, reaching 98.07% in the feces of C57BL/6J mice. The relative abundance of B. pseudolongum in feces was higher than that in cecal contents. In the whole intestinal microbiota, the relative abundance of B. pseudolongum was 9.31% and 9.49% in cecal contents and feces of C57BL/6J mice, respectively, and 0.72% and 1.64% in BALB/c mice, 3.29% and 4.36% in ICR mice, and 0.75% and 2.32% in KM mice. Meanwhile, the relative abundance of B. longum increased and that of B. pseudocatenulatum decreased slightly in both feces and fecal contents. Thus, the intestinal microbiota of the four genotypes of mice showed the same changes after oral administration of FOS, both in feces and cecal contents. FOS stimulated the proliferation of B. pseudolongum in the intestinal microbiota of the four genotypes of mice, and the proliferation was more remarkable in C57BL/6J mice.
Fig. 5.
Effects of FOS on the relative abundance of Bifidobacterium species in the Bifidobacterium genus in the feces (A) and cecal contents (B), and the relative abundance of Bifidobacterium species in the intestinal microbiota (C).
3.7. Changes in gut microbial functions predicted via PICRUSt2
PICRUSt2 was used to perform functional predictions of 16S rDNA sequencing results (Fig. 6). Based on the KEGG pathway database, third-level analysis of functional categories of the KEGG pathway suggested that the relative abundance of most predicted functional categories changed significantly among the different groups (Kruskal–Wallis test, p < 0.05). Compared with the control group, 29 functional modules (14 increased) were significantly altered in the feces of the C57BL/6J mice. The majority of the gene families were involved in membrane transport, replication and repair, amino acid metabolism, and carbohydrate metabolism. FOS improved replication and repair, translation, amino acid metabolism, energy metabolism, nucleotide metabolism, and cofactor and vitamin metabolism in C57BL/6J mice. FOS significantly promoted bacterial metabolism and cellular processes in the feces of BALB/c mice and membrane transport in the cecal contents of BALB/c mice. Similarly, in the feces and cecal contents of KM mice, FOS mainly increased the bacterial metabolism, cellular processes, and replication and repair. Level 2 KEGG pathways associated with “chromosome”, “pyruvate metabolism”, “phenylalanine, tyrosine and tryptophan biosynthesis” and “valine, leucine and isoleucine biosynthesis” were relatively the most abundant in the feces of ICR mice, while chloroalkane and chloroalkene degradation were predicted to be more abundant in the cecal contents.
Fig. 6.
Effect of FOS on intestinal microbiota functions predicted by PICRUST2.
3.8. Identification of B. pseudolongum in mouse feces
To compare the ability of B. pseudolongum from mice of different genotypes to utilize FOS, eight strains of B. pseudolongum species were screened from mouse feces, including C1 and C2 from C57BL/6J mice, B3 and B10 from BALB/c mice, I5 and I6 from ICR mice, and K1 and K2 from KM mice (Table 3). The OD600 and pH values were measured after culturing in GAM medium. The colonies of all strains turned yellow in a culture medium supplemented with bromocresol violet as indicators and FOS as the sole carbon source, indicating that all screened strains could produce acid during FOS fermentation. The OD600 and pH were measured after 24 h of culture in GAM medium. The pH of the medium was approximately 4.7 for all strains. The OD600 of strains C1 and I6 was the highest, reaching 2.19 ± 0.08 and 2.15 ± 0.28, respectively. The results indicated a difference in FOS utilization among the strains.
Table 3.
Strains of B. pseudolongum isolated from mouse feces.
| Origin | Strains | OD600 | pH | FOS consumption |
|---|---|---|---|---|
| C57BL/6J mice | C1 | 2.19 ± 0.08 | 4.73 ± 0.04 | + |
| C2 | 1.88 ± 0.45 | 4.76 ± 0.01 | + | |
| Balb/C mice | B3 | 1.90 ± 0.36 | 4.76 ± 0.02 | + |
| B10 | 1.91 ± 0.51 | 4.77 ± 0.04 | + | |
| ICR mice | I5 | 1.82 ± 0.40 | 4.76 ± 0.01 | + |
| I6 | 2.15 ± 0.28 | 4.75 ± 0.01 | + | |
| KM mice | K1 | 1.92 ± 0.39 | 4.72 ± 0.04 | + |
| K2 | 1.78 ± 0.57 | 4.72 ± 0.01 | + |
Notes: The OD600 and pH of the B. pseudolongum strains after incubation in the GAM medium showed no significant differences (ANOVA, p > 0.05).
3.9. Differences in FOS utilization during bacterial growth
To further determine the differences in FOS utilization among the strains during bacterial growth, we monitored the growth of the identified species in GAM medium with FOS as the sole carbon source (Fig. 7). The strain C1 from C57BL/6J mice and I6 from ICR mice grew faster and reached a plateau in approximately 9 h. The strains from BALB/c and KM mice had similar growth profiles and showed longer generation times. We measured the number of viable bacteria at the end of the logarithmic phase. The maximum viable count of C1 reached 6.60 × 108 CFU/mL at the stationary phase, and no significant difference was observed between the abundance of C2 and I6 (over 4.00 × 108 CFU/mL), whereas the number of the other strains showed fewer viable counts. The medium supernatants from 0 h, initial logarithmic phase, end of the logarithmic phase, and 24 h incubations were collected for the detection of FOS content. It was evident from HPLC that all strains utilized GF2 and GF3 to varying degrees after incubation, confirming the significant differences in their ability to utilize FOS (see Fig. 8).
Fig. 7.
The growth curve of the strains in GAM medium with FOS as the sole carbon source.
Fig. 8.
Contents of FOS in the culture supernatant of the isolated bacteria after 0 h, initial logarithmic phase, the end of logarithmic phase and 24 h incubation. The retention times of GF2, GF3 and GF4 were 16.65, 19.02 and 21.03 min, respectively.
The rate of FOS consumption in the logarithmic phase of the screened strains was calculated by determining the FOS content in the early and end logarithmic phases, as shown in Fig. 9. None of the eight screened strains utilized GF4, and significant differences were observed in the utilization rates of GF2 and GF3. The strains C1 and C2 isolated from C57BL/6J mice could not utilize GF3, but the two strains utilized GF2 at the highest rate of 0.89 ± 0.18 g/h and 0.50 ± 0.09 g/h, respectively. I5 utilized FOS at a rate similar to C2 but utilized GF3 at the highest rate among the eight strains (0.07 ± 0.02 g/h). The strains originating from BALB/c and KM mice utilized GF2 at a lower rate of approximately 0.30 g/h. Thus, the ability of B. pseudolongum strains obtained from different genotypes of mice to utilize FOS differed significantly. The two strains of B. pseudolongum derived from C57BL/6J mice exhibited a shorter growth generation and utilized FOS at the fastest rate, whereas the strains derived from BALB/c and KM mice had a lower ability to utilize FOS. Combined with the previous results of this study, we hypothesized that FOS utilization capacity might be one of the key factors to influence the response of B. pseudolongum to FOS in vivo.
Fig. 9.
The rates of FOS consumption during the logarithmic phase.
4. Discussion
Few previous studies on diet and intestinal microflora have compared the response of intestinal microflora in mice with different genotypes to diet in the same experiment. Our previous studies showed that FOS significantly promoted the proliferation of B. pseudolongum in C57BL/6J mice (Mao et al., 2018; Gu et al., 2019, Gu et al., 2022a). Considering that differences in the background strains in mice with different genotypes affect the response of bacteria species to FOS, we analyzed the intestinal microbiota of the four commonly used strains of mice before and after FOS ingestion in this study. The results obtained are helpful for the selection of suitable strains of mice and for the detection of the mechanism of the response of B. pseudolongum to FOS.
Based on 16S rDNA sequencing, we detected intestinal background bacteria strains in mice with different genotypes. The intestinal microbiota of the four genotypes exhibited similarities in composition and differences in relative abundance. The differences might be because the genotype accounts for a substantial fraction of the abundance of the most common microbiota in a certain environment (Org et al., 2015). Muribaculaceae occupied a high relative abundance in mice, especially in the feces of BALB/c mice (over 40%). Muribaculaceae (belonging to the genus Bacteroides) is a common and abundant inhabitant of mammalian intestines, especially in mice. Muribaculaceae specializes in polysaccharide fermentation to primarily produce propionate (Ormerod et al., 2016; Smith et al., 2021). Lachnospriaceae and Alistipes showed higher relative abundances in KM and ICR mice, respectively. Jiang et al. (2020) and Chen et al. (2020) also reported the highest abundances of Lachnospriaceae and Muribaculaceae in KM mice. Lachnospriaceae is a butyric-acid-producing bacterium that is negatively correlated with intestinal disorders and other diseases (Flint et al., 2012).
Moreover, the relative abundance of cecal microbes also differed considerably from that of fecal microbes, which differentiated functional and metabolic differences between the cecal and fecal microbiota in mouse (Tanca et al., 2017). The variation tend in the relative abundance of fecal and cecal microbes in this study were similar to that in our previous study. We previously analyzed the taxonomic composition of the intestinal microbiota at different sampling sites in C57BL/6J mice and described the composition of the intestinal microbiota along the intestinal tract (Gu et al., 2019). Physicochemical conditions (pH, redox potential, oxygen, and salts concentration) and biochemical interactions (with molecules released by the intestinal epithelium, immune cells, and other microbes) change along the gastrointestinal tract, that might shape the microbiota (Haange et al., 2012).
The intestinal microbiota of inbred and closed colony mice was consistently shaped by FOS, indicating that diet played a dominant role in shaping the gut microbial ecology. The effect of FOS on the intestinal microbiota varied with different genotypes despite the similar commensal composition of the mice. The initial relative abundance of Bifidobacterium was similar (<0.10%) among the four genotypes of mice but increased to varying degrees after FOS ingestion (Fig. 4), especially in C57BL/6J mice, which increased to over 10% in this study. The differences in the response of intestinal microbiota to FOS under specific feeding conditions might be related to the host genotype. Bonder et al. (2016) provided evidence of a gene-diet interaction in the regulation of Bifidobacterium abundance. The abundance of taxa regulated by the host genotype is associated with diet sensing, metabolism, and immunity (Goodrich et al., 2016). Khachatryan et al. also proposed the importance of host genetics in the determination of host-microbe interactions, but the exact mechanisms underlying this interaction are not fully understood (Khachatryan et al., 2008).
B. pseudolongum is a widely distributed gut commensal in animals and is more likely to colonize the gut of mice (Lugli et al., 2019; Xiao et al., 2021). B. pseudolongum dominated the genus Bifidobacterium and was significantly promoted by FOS in the feces and cecum of mice of different genotypes. This effect was more pronounced in C57BL/6J mice. In our previous study, we investigated the ability of several FOS-metabolizing species to utilize FOS and found that B. pseudolongum utilized FOS with a shorter growth generation time, faster rate of FOS utilization, and higher enzymatic activity, which was presumed to be one of the mechanisms for the competitive advantage of B. pseudolongum in vivo (Gu et al., 2022b). Hopkins et al. (1998) also concluded that B. pseudolongum exhibited a higher growth rate and bacterial cell yield than B. longum and B. breve when cultured in a medium with FOS as the sole carbon. In this study, B. pseudolongum was stimulated differently in different strains of mice, which might also be due to the difference in the ability of B. pseudolongum to utilize FOS in different strains of mice.
To determine the relationship between B. pseudolongum competition and FOS in mice with different genetic backgrounds, we isolated eight strains of B. pseudolongum from the feces of the four genotypes of mice and examined their ability to utilize FOS. The main components of FOS are GF2 (60.60%) and GF3 (32.45%), with one more fructose unit than GF2. According to our previous study (Gu et al., 2021), B. pseudolongum translocated FOS to intracellular hydrolysis by permease, and the transport capacity might decrease as the degree of poly-merization increases. Therefore, the B. pseudolongum strains isolated from different genotypes of mice showed a lower rate of GF3 utilization and a higher utilization rate of GF2. Meanwhile, we found differences in the utilization of FOS by strains from different sources, and the strains derived from the same feces also showed significant differences in utilization capacity in vitro, suggesting the importance of analyzing bacterial function at the strain level. The fermentation capabilities of carbohydrates vary considerably among B. pseudolongum strains (Lugli et al., 2019). The rate of FOS utilization by different strains might be related to the efficiency of the transport system and the enzymatic activity of the hydrolase. This study presents for the first time the dramatic differences in the ability and pattern of FOS utilization by B. pseudolongum strains, and further genomic characterization of the eight strains is necessary.
The two strains of B. pseudolongum screened from the feces of C57BL/6J mice were more efficient in utilizing FOS, which is consistent with the dramatic increase in C57BL/6J mice, indicating that stronger utilization of FOS by B. pseudolongum strains might determine an advantage to compete in vivo. Efficient (arabino) xylan utilization contributes to Prevotella dominance in the human gut (Gálvez et al., 2020). Thus, dietary interventions are a potential tool to modulate the gut microbiota and further impact host health (Kovatcheva-Datchary and Arora, 2013). Future studies on the mechanism of the bacterial response to diet in vivo would provide crucial clues for understanding the relationship between intestinal microflora and health.
5. Conclusion
In this study, we analyzed the intestinal microbiota of four commonly used mice, C57BL/6J, BALB/c, ICR, and KM, before and after FOS intervention. The intestinal microflora composition of the mice with different genotypes was similar but differed in relative abundance. FOS promoted the proliferation of B. pseudolongum in all four genotypes of mice, with the most significant effect on C57BL/6J mice. The ability and manner of FOS utilization by B. pseudolongum strains from different genotypes of mice varied greatly, with strains derived from C57BL/6J mice being more capable of utilizing FOS. The gut microbial analysis of mice with different genotypes complemented our previous studies. These findings provided information on the gut background bacteria strains for experiments related to the intestinal microbiota to facilitate the selection of murine models and suggested a possible correlation between FOS utilization ability and the response of bacteria strains to FOS in vivo. Further studies on the ability of bacteria strains to utilize FOS and competition in the intestine will help us to better understand the mechanisms of the intestinal microbial response to diet.
CRediT authorship contribution statement
Jiayu Gu: Writing – original draft, preparation, Investigation, Data curation, Formal analysis. Shumao Cui: Project administration, Funding acquisition. Xin Tang: Project administration, Methodology. Zhenmin Liu: Writing – review & editing, Conceptualization, Supervision. Jianxin Zhao: Writing – review & editing, Project administration. Hao Zhang: Conceptualization, Supervision. Bingyong Mao: Writing – original draft, preparation, Writing – review & editing, Conceptualization, Methodology, Funding acquisition. Wei Chen: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 31972086, 32172173, 32072197), Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province, and Postgraduate Research & Practice Innovation Program of Jiangsu Province.
Data availability
Data will be made available on request.
References
- Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F., Fisher E.M. Genealogies of mouse inbred strains. Nat. Genet. 2000;24(1):23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Bonder M., Kurilshikov A., Tigchelaar E., Mujagic Z., Imhann F., Vila A.V., Deelen P., Vatanen T., Schirmer M., Smeekens S.P., Zhernakova D.V., Jankipersadsing S.A., Jaeger M., Oosting M., Cenit M.C., Masclee Ad A.M., Swertz M.A., Li Y., Kumar V., Joosten L., Harmsen H., Weersma R.K., Franke L., Hofker M.H., Xavier R.J., Jonkers D., Netea M.G., Wijmenga C., Fu J., Zhernakova A. The effect of host genetics on the gut microbiome. Nat. Genet. 2016;48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- Carmody R.N., Gerber G.K., Luevano J.M., Jr., Gatti D.M., Somes L., Svenson K.L., Turnbaugh P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17(1):72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Zhou S.S., Li J.W., Huang X.Y., Cheng J.H., Jiang X.J., Qin W., Liu Y.T., Liu A.P., Zhang Q., Lin D.R., Zhang Z.Q., Chen D.W. Xyloglucan compounded inulin or arabinoxylan against glycometabolism disorder via different metabolic pathways: gut microbiota and bile acid receptor effects. J. Funct.Foods. 2020;74 [Google Scholar]
- Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut microbe. 2012;3(4):289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez E.J.C., Iljazovic A., Amend L., Lesker T.R., Renault T., Thiemann S., Hao L.X., Roy U., Gronow A., Charpentier E., Strowig T. Distinct polysaccharide utilization determines interspecies competition between intestinal Prevotella spp. Cell Host Microbe. 2020;28:838–852. doi: 10.1016/j.chom.2020.09.012. [DOI] [PubMed] [Google Scholar]
- Goodrich J.K., Davenport E.R., Waters J.L., Clark A.G., Ley R.E. Cross-species comparisons of host genetic associations with the microbiome. Science. 2016;352(6285):532–535. doi: 10.1126/science.aad9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T., Spector T.D., Clark A.G., Ley R.E. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J.Y., Cui S.M., Tang X., Liu Z.M., Zhao J.X., Zhang H., Mao B.Y., Chen W. Effects of fructooligosaccharides (FOS) on the composition of cecal and fecal microbiota and the quantitative detection of FOS-metabolizing bacteria using species-specific primers. J. Sci. Food Agr. Online. 2022 doi: 10.1002/jsfa.11884. [DOI] [PubMed] [Google Scholar]
- Gu J.Y., Mao B.Y., Cui S.M., Liu Z.M., Zhang H., Zhao J.X., Chen W. Metagenomic insights into the effects of fructooligosaccharides (FOS) on the composition of luminal and mucosal microbiota in C57BL/6J mice, especially the Bifidobacterium composition. Nutrients. 2019;11:2431–2443. doi: 10.3390/nu11102431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J.Y., Mao B.Y., Cui S.M., Tang X., Liu Z.M., Zhao J.X., Zhang H. Bifidobacteria exhibited stronger ability to utilize fructooligosaccharides, compared with other bacteria in the mouse intestine. J. Sci. Food Agric. 2022;102:2413–2423. doi: 10.1002/jsfa.11580. [DOI] [PubMed] [Google Scholar]
- Haange S.B., Oberbach A., Schlichting N., Hugenholtz F., Smidt H., Von Bergen M., et al. Metaproteome analysis and molecular genetics of rat intestinal microbiota reveals section and localization resolved species distribution and enzymatic functionalities. J. Proteome Res. 2012;11:5406–5417. doi: 10.1021/pr3006364. [DOI] [PubMed] [Google Scholar]
- Hildebrand F., Nguyen T.L., Brinkman B., Yunta R.G., Cauwe B., Vandenabeele P., Liston A., Raes J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14(1):R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M.J., Cummings J.H., Macfarlane G.T. Inter-species differences in maximum specific growth rates and cell yields of bifidobacteria cultured on oligosaccharides and other simple carbohydrate sources. J. Appl. Microbiol. 1998;85:381–386. [Google Scholar]
- Hugenholtz F., de Vos W.M. Mouse models for human intestinal microbiota research: a critical evaluation. Cell. Mol. Life Sci. 2018;75:149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X.Y., Ding H.Y., Liu Q., Wei Y.X., Zhang Y.J., Wang Y.P., Lu Y.Q., Ma A.G., Li Z.C., Hu Y.F. Effects of peanut meal extracts fermented by Bacillus natto on the growth performance, learning and memory skills and gut microbiota modulation in mice. Br. J. Nutr. 2020;123(4):383–393. doi: 10.1017/S0007114519002988. [DOI] [PubMed] [Google Scholar]
- Khachatryan Z.A., Ktsoyan Z.A., Manukyan G.P., Kelly D., Ghazaryan K.A., Aminov R.I. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs A., Ben-Jacob N., Tayem H., Halperin E., Iraqi F.A., Gophna U. Genotype is a stronger determinant than sex of the mouse gut microbiota. Microb. Ecol. 2011;61:423–428. doi: 10.1007/s00248-010-9787-2. [DOI] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P., Arora T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 2013;27:59–72. doi: 10.1016/j.bpg.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Lugli G.A., Duranti S., Albert K., Mancabelli L., Napoli S., Viappiani A., Anzalone R., Longhi G., Milani C., Turroni F. Unveiling Genomic diversity among members of the species Bifidobacterium pseudolongum, a widely distributed gut commensal of the animal kingdom. Appl. Environ. Microbiol. 2019;85 doi: 10.1128/AEM.03065-18. e03065-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B.Y., Gu J.Y., Li D.Y., Cui S.M., Zhao J.X., Zhang H., Chen W. Effects of different doses of fructooligosaccharides (FOS) on the composition of mice fecal microbiota, especially the Bifidobacterium composition. Nutrients. 2018;10:1105–1119. doi: 10.3390/nu10081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B.Y., Li D.Y., Zhao J.X., Ai C.Q., Zhang H., Chen W. Lactulose differently modulates the composition of luminal and mucosal microbiota in C57BL/6J mice. J. Agric. Food Chem. 2016;64:6240–6247. doi: 10.1021/acs.jafc.6b02305. [DOI] [PubMed] [Google Scholar]
- Org E., Parks B.W., Joo J.W., Emert B., Schwartzman W., Kang E.Y., Mehrabian M., Pan C., Knight R., Gunsalus R., Drake T.A., Eskin E., Lusis A.J. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015;25:1558–1569. doi: 10.1101/gr.194118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormerod K.L., Wood D.L.A., Lachner N., Gellatly S.L., Daly J.N., Parsons J.D., Dal'Molin C.G.O., Palfreyman R.W., Nielsen L.K., Cooper M.A., Morrison M., Hansbro P.M., Hugenholtz P. Genomic characterization of the uncultured bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4:36. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks B.W., Nam E., Org E., Kostem E., Norheim F., Hui S.T., Pan C., Civelek M., Rau C.D., Bennett B.J., Mehrabian M., Ursell L.K., He A., Castellani L.W., Zinker B., Kirby M., Drake T.A., Drevon C.A., Knight R., Gargalovic P., Kirchgessner T., Eskin E., Lusis A.J. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metabol. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangeetha P.T., Ramesh M.N., Prapulla S.G. Recent trends in the microbial production, analysis and application of fructooligosaccharides. Trends Food Sci. Technol. 2005;16:442–457. [Google Scholar]
- Smith B.J., Miller R.A., Schmidt T.M. Muribaculaceae genomes assembled from metagenomes suggest genetic drivers of differential response to acarbose treatment in mice. Am. Soc. Microbiol. News. 2021;6(6) doi: 10.1128/msphere.00851-21. e00851-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spor A., Koren O., Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Tanca A., Manghina V., Fraumene C., Palomba A., Abbondio M., Deligios M., Silverman M., Uzzau S. Metaproteogenomics reveals taxonomic and functional changes between cecal and fecal microbiota in mouse. Front. Microbiol. 2017;8:391. doi: 10.3389/fmicb.2017.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.L., Hu L.J., Yan S., Jiang T., Fang S.G., Wang G., Zhao J.X., Zhang H., Chen W. Effects of different oligosaccharides at various dosages on the composition of gut microbiota and short-chain fatty acids in mice with constipation. Food Funct. 2017;8:1966–1978. doi: 10.1039/c7fo00031f. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Zhao J.X., Zhang H., Zhai Q.X., Chen W. Colonized niche, evolution and function signatures of Bifidobacterium pseudolongum within Bifidobacterial genus. Foods. 2021;10(10):2284. doi: 10.3390/foods10102284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang J.N., Sun B.H., Liu Q., Ma J., Zhang Q., Liu Y.X., Chen N., Chen F. The role of genotype and diet in shaping gut microbiome in a genetic vitamin A deficient mouse model. J. Genet. Genomics. 2022;49(2):155–164. doi: 10.1016/j.jgg.2021.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.